Experimental Protocol for Sedimentation Equilibrium Analytical Ultracentrifugation
Andrea Balbo, Patrick Brown, and Peter Schuck
Last updated in July 2007
This step-by-step protocol illustrates a sedimentation equilibrium (SE) study of a heterogeneous interaction between two proteins ‘A’ and ‘B’ forming a reversible complex. The goal of the experiment is the determination of the binding constant and the binding stoichiometry. It includes the determination of the molar mass of a non-interacting protein (or stable protein complex), and can be easily adapted to the study of protein self-association. Again, the details of the practical steps are given in conjunction with the workflow of the data analysis, which is closely tied to the experimental configuration. It will be assumed that a sedimentation velocity (SV) study of the same proteins and their mixture has taken place before, for example, using the SV protocol provided by us.
It is essential that the reader be familiar with the document "general introduction to sedimentation equilibrium", which explains the conceptual and theoretical aspects, as well as the requirements with regard to sample preparation, purity, and amounts. The general introduction to sedimentation equilibrium also outlines the problem and strategies for the mathematical data analysis, which is a consideration indispensable for the successful experimental setup. Modern analysis techniques gain substantially in information and precision from fitting several experimental data sets globally. Since this introduces an increasing number of fitting parameters, it is imperative to take advantage of available constraints, such as mass conservation and relationships between the loading concentrations from different sample solutions, which can be generated by appropriate experimental designs. This is described in detail in Vistica et al.
The protocols have been written here in an effort to make the experiment most informative. By following the protocol, one is able to generate several parameter constraints making the analysis most manageable. Much care and foresight should go into the planning, preparation, and data acquisition stages, to minimize errors and noise. Also, in the design and execution of the experiment it is important to already anticipate the later data analysis stage, in order to obtain reliable, high quality data with sufficient information to determine the thermodynamic parameters precisely.
Hands-on training in the techniques described here can be obtained from the Workshop on Hydrodynamic and Thermodynamic Analysis of Macromolecules with SEDFIT and SEDPHAT at the National Institutes of Health, which takes place twice a year in Bethesda, Maryland.
* Analytical Ultracentrifuge (Beckman Coulter) equipped with absorption optical scanner (ABS) and optionally Rayleigh laser interferometer (IF) imaging system
* Spectrophotometer (preferably dual-beam, any supplier) may be required for the determination of the protein concentration via UV absorbance
* Densitometer (Anton Paar) may be required for measuring the solvent density if non-tabulated buffer components are used (tabulated values are available for most common buffers and salts)
* Computer (PC) equipped with software to perform the data analysis (this should not be a PC simultaneously running the ultracentrifugal data acquisition to avoid interference):
(1) SEDFIT and SEDPHAT for transforming and analyzing the sedimentation equilibrium data;
(2) SEDNTERP for calculating the protein partial-specific volumes and extinction properties, as well as buffer density;
(3) general purpose software to store screenshots for documentation (MS Word, powerpoint or equivalent)
(4) internet connection to consult the SEDPHAT getting started tutorials, the step-by-step tutorials for sedimentation equilibrium analysis, the command reference manual, and the SEDFIT/SEDPHAT search bar. For an introductory overview, point your web-browser to http://www.analyticalultracentrifugation.com/sedphat/sedimentation_equilibrium_for_interacting_systems.htm
1. Collect all prior knowledge. Although not all of this information is essential, and some may not be possible or fascile to obtain, it can often be helpful during and after an analysis. It helps to have this information in the planning stages for determining rotor speeds, time to equilibrium, etc.; also during the analysis stage as far as inputting starting guesses for the fitted parameters, and after an analysis to determine if fitted parameters make sense.
a. Obtain the amino acid sequence(s) of the analytes. (Essential)
The molar mass
as well as the extinction coefficient at 280 nm (and 250 nm) and the ![]() values can usually be calculated from a known amino acid
sequence (via SEDNTERP). For proteins that are known not to self-associate, the
buoyant molar mass (essentially the
values can usually be calculated from a known amino acid
sequence (via SEDNTERP). For proteins that are known not to self-associate, the
buoyant molar mass (essentially the ![]() ) values of the individual proteins will be measured in this
protocol, but it is useful to compare this value with the theoretical sequence
value. Also, inquire about whether any free cysteines are known to be present
as these can lead to irreversible crosslinking.
) values of the individual proteins will be measured in this
protocol, but it is useful to compare this value with the theoretical sequence
value. Also, inquire about whether any free cysteines are known to be present
as these can lead to irreversible crosslinking.
b. Obtain mass spectrum. (Optional, but sometimes essential)
Ensure agreement between mass calculated from composition with that obtained by mass spectrometry. Some possible explanations for disagreement include: proteolytic cleavage, post-translational modification, irreversible aggregation, etc. For glycoproteins, the determination of the carbohydrate mass fraction by mass spectrometry is desirable, in order to calculate a good estimate of the partial-specific volume of the glycoprotein.
c. Observe size-exclusion chromatogram. (Highly desirable)
Knowing where a protein of specific molecular weight elutes can give insight into the asymmetry of the protein (not important in SE but is in SV) as well as the possible heterogeneity of the collected fractions. Even more importantly, size-exclusion chromatography should be the last purification step, if at all possible (see below).
d. Observe a PAGE gel image. (Essential)
Native gels and denaturing gels under reducing/non-reducing conditions can be useful in estimating protein mass, and in observing the presence of higher-order complexes and/or crosslinked aggregates.
e. Understand the composition of the buffer. (Essential)
Useful information may be how the buffer was obtained, whether it is filtered dialysate or flow-through of the SEC column. Are there any additives that can be observed with the optical detection system which will contribute to the shape of the gradient or that will affect the manner in which the protein sediments and diffuses. Measure or calculate buffer density and viscosity for the temperature at which the SE experiment will be performed. See below section I.3.
f. Other information: (Optional)
Knowledge of the stability of the protein is important for designing the experiment. If the protein is prone to degradation or aggregation at room temperature it may be more desirable to acquire data at lower temperatures or for shorter time periods (using shorter solution columns or at fewer rotor speeds). Additionally, this information may be useful in the analysis stage wherein inclusion of proteolytic fractions or discrete aggregates can be incorporated into the model.
It can be useful to consider results from other biophysical experiments that have been performed on the system of interest (or similar systems) as a guide to reasonable estimates of affinity or stoichiometry in the planning and/or analysis stages.
It may be helpful from a troubleshooting perspective to understand the expression system, and steps taken in the purification process.
2. Conduct a SV experiment in order to assess the purity of the sample and to gain some qualitative assessment of the concentration dependence of the association state. Generally, purity of better than 95% is required for SE. Small molar mass impurities (e.g. with masses in the range of a few percent of the proteins of interest), which frequently go undetected by SDS-PAGE, will complicate the analysis and should be removed, if possible. Irreversible small oligomeric aggregates will generally also limit the precision of the binding parameters to be measured. Therefore, size-exclusion chromatography is highly recommended as the last preparative step. Contamination with very large aggregates, distinctly larger than the molar mass of the complex, can usually be tolerated. The SV run should be conducted at a wide range of sample concentrations, such that it will provide insight into the oligomeric state of each protein in solution and the detection of reversible self-association of the two components. It will also permit crude estimation of the binding constant (KD* – e.g., an empirical estimate of the concentration at which the AB complex is half-saturated). This will greatly help in planning the sample concentrations and the rotor speeds for the SE experiment.
3.
Consider the buffer
composition. Phosphate buffers are transparent in the far UV and
are suitable for both ABS and IF detection. For example, PBS (phosphate
buffered saline) will work well. When using other buffers, see Table 1 and Table 2. If the buffer
conditions and the protein extinction do not govern the choice of the detection
system, use the IF system. Make sure you know all the components of the
buffer solution being used. Then, measure the density and viscosity of the
buffer, at the actual AUC run temperatures, with the densitometer and
viscosimeter, respectively, or calculate them with SEDNTERP. (The viscosity is
strictly required only for SV experiments, but can be useful when using
additives, such as glycerol or sucrose, in SE that can increase the time
required to attain equilibrium.) As a preliminary step, it might be necessary
to establish sedimentation/optical properties of the buffer before analyzing the
actual proteins, if, for example, detergents or other sedimenting buffer
components are present at concentrations leading either to signal gradients or
density gradients. Buffer conditions are slightly less restrictive for SE with
regard to components that can create density gradients at the higher rotor
speeds of SV: For example, a small percentage of glycerol can be tolerable in
SE using the ABS optics, provided the potential effect on ![]() from preferential protein hydration and the extended
equilibration time is taken into account. As in the SV experiments (Step
I.2.), a chemically identical reference buffer is required when using IF
optics. Measure the buffer density or use SEDNTERP to calculate the appropriate
value.
from preferential protein hydration and the extended
equilibration time is taken into account. As in the SV experiments (Step
I.2.), a chemically identical reference buffer is required when using IF
optics. Measure the buffer density or use SEDNTERP to calculate the appropriate
value.
4. Choose the rotor and centerpieces. The 8-hole rotor and double sector centerpieces are the recommended option.
The double sector centerpieces allow for longer solution columns than the 6-channel centerpieces. While requiring longer time to reach equilibrium, longer solutions columns provide for increased curvature and more data points in the gradient which leads to better precision in the analysis. When 6-channel centerpieces cannot be avoided due to the large number of samples that need to be run, fill them with the maximum volume (130 – 140 microliters).
The 8-hole rotor allows for at least twice as many samples to be analyzed in one experiment than by using the 4-hole rotor. If using an 8-hole rotor, reserve one cell for each of the individual proteins to run as a control, and use the 5 (or 6) remaining cells for mixtures. With a 4-hole rotor, consider using three 6-channel centerpieces to increase the number of samples. If these options are not available and the individual components have to be studied in a separate run, note that small differences in the purity or presence of irreversible aggregates from different preparations or from different time-points can be confused with and correlate with the detection of putative heterogeneous complexes. In practice, this can be a problem, particularly for weak interactions.
5. Select approximate sample concentrations to span a range that will generate different relative populations of free and complex species: For hetero-associations with unknown stoichiometry, it can be advantageous to choose a constant concentration of ‘A’ 2fold to 5fold greater than the estimated KD*, and to titrate constant ‘A’ in different samples with variable amounts of ‘B’ to generate a range of molar ratios comprising the suspected possible stoichiometries. (The constancy of loaded amounts of ‘A’ can later serve as a powerful constraint on the data analysis.) This may be combined with a second set of A and B at the same molar ratios used initially, but at a concentration of ‘A’ several-fold below KD*. In contrast, if the stoichiometry is known, it can be advantageous to make a stock mixture at a concentration of > 10fold KD* with a molar ratio equal to the complex stoichiometry, and to generate the different samples by serial dilution to span a large range of concentrations. (Here, the constancy of the molar ratio generates a useful constraint for the data analysis.) If the available maximal sample concentrations are too low (e.g. only one tenth of KD*) adjust the following protocol by using a longer solution column, tolerating the longer equilibration time but taking advantage of the concentrating effect of the centrifugal field. The dynamic range of the optical system (see Table 1) must be considered for selecting sample concentrations.
6. Choose the optical detection and double-check the sample concentrations with regard to optical detectability. The key for selecting the optical system is to generate SE profiles in such a way that the contributions from free species, which attain relatively shallower exponential distributions, and complex species, which generate relatively steeper exponentials, can all be detected and distinguished (see the introduction to SE): Proteins vary greatly in the number of tyrosine and tryptophan residues, and therefore in their extinction spectrum in the far UV. Because the ABS system can take advantage of this difference to help distinguish between the components ‘A’ and ‘B’, ABS with multi-wavelength analysis is usually preferred. Obviously, this technique will be more powerful for protein/nucleic acid interactions, or if one of the interacting components has an intrinsic chromophore in the near UV or VIS or if an extrinsic label can be attached. Attaching an extrinsic label is frequently very useful if the molar masses of ‘A’ and ‘B’ differ by less than 20% or more than 80%. However, in many cases this may not be necessary when SE data are obtained at multiple rotor speeds in conjunction with mass conservation analysis (a technique described in the following).
For the choice of the protein concentrations for ABS detection, if consistent with the order of magnitude of KD*, and the considerations described in Step I.5., maximize the dynamic range of the ABS system at the wavelengths of 230 nm, 250 nm, and 280 nm: This can be achieved in samples with total absorbances of 0.1 OD230, 0.2 OD280, 0.5 OD280, and 0.5 OD250. Note that the centrifugal field will create a gradient locally decreasing or increasing the concentrations several-fold. Higher concentrations can be used, for example, with very high KD*, but this may require the use of the IF optics to not be constrained with the limited linearity of the ABS optics. For the two individual protein samples included as controls in the same run, choose a concentration of 0.5 OD280. This will permit the determination of their extinction coefficients at 230 and 250 nm.
The use of the ABS system may be prohibited by the solvent conditions, or it may not provide a sufficient dynamic range of protein concentrations for the detection of weak interactions. Also, one or both of the proteins under study may not have a sufficiently high extinction coefficient to generate signals > 0.05 OD at any or all of the absorbance wavelengths permitted by the buffer system. In this case, the IF system can be used, with the considerations for sample concentrations as outlined above (Step I.5.).
Maximum information can be gained if the IF system is used in conjunction with the ABS system, which improves the distinction between the two proteins in a multi-signal analysis. Note that the use of the IF system for SE will require the mechanical stabilization of the cell assembly (Step II.2a.). In the combination of IF with ABS data, in particular if the solution volumes and rotor speeds are used as suggested in this protocol, the radial-dependent baseline can be computationally determined and water blanks are not essential.
7. The choice of the sample volume and rotor temperature is dictated mainly by the protein size and stability. Except when studying interacting systems with very slow chemical rates (frequently encountered with very high affinity systems), the sedimentation time is governed by the diffusion of the proteins throughout the solution column. As a standard choice, use 170 microliter samples and expect equilibration at the first speed within 48 hours (usually less time is required at the following, sequentially higher rotor speeds). True attainment of sedimentation equilibrium must be checked experimentally (see below). To avoid longer equilibration times required for larger protein complexes > 200 kDa, usually the sample volume is decreased to 140 – 150 microliters. For samples that do not possess sufficient stability for attaining equilibrium at three rotor speeds, 100 – 120 microliter samples can be used. (Alternatively, shorter column techniques are available, but they only provide information on the approximate average molecular weight as a function of loading concentration, which does not provide as much detail as the long-column technique and requires a different analytical strategy than described in this protocol [see, e.g., molecular weight isotherm analysis in SEDPHAT].) On the other hand, when studying small proteins (e.g., < 5 kDa) or peptides, larger solution columns of up to 400 microliter may be advantageous to generate equilibrium gradients with sufficient curvature at the accessible rotor speeds. In this case, the higher diffusion coefficient of smaller species leads to an only moderate increase in equilibration time.
To shorten the time to attain equilibrium, an initial overspeeding period for a few hours at three-fold the first equilibrium rotor speed can be used, but the concentration profiles should be monitored so as not to exceed a value two or three times the loading concentration at the bottom of the cell (this is described in Step III.3. below).
Usually, the rotor temperature is an important factor for the stability, and unless thermodynamic considerations prescribe the use of a particular temperature (the binding constant is temperature dependent following RT ln(KA) = DG = DH – TDS), low temperatures such as 4 – 8 °C are recommended.
8. The rotor speeds must be selected to generate concentration gradients with shapes that include both shallow and steeper regions. It is highly useful to acquire data sequentially at multiple rotor speeds, generating first relatively shallow gradients with ratios of bottom concentrations to meniscus concentrations of 3:1 – 5:1, and finally, at the highest rotor speed, a condition where the meniscus region is free of protein, termed ‘meniscus depletion’. The shapes of typical profiles are like this:
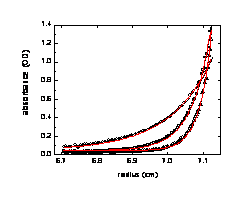
For 170 – 180 microliter
samples, select the three rotor speeds according to the formula
w1 = 0.75´105/M1/2,
w2 = 1.2´105/M1/2,
w3 = 1.5´105/M1/2,
inserting for M the average molar masses (in kDa) of the components and
the expected complex. (For samples of unknown mass, these values can be
estimated from the previous SV experiment, or from the initial slopes in an
Archibald analysis of the early experimental SE data close to the meniscus and
bottom (see Schuck & Millar 1998),
available in SEDFIT.) For different sample volumes, measure the new column
height l from initial scans and use the formula ![]() to correct the value from those given above. A slightly
higher value for the highest rotor speed generating steeper gradients can be
applied when using the IF optics.
to correct the value from those given above. A slightly
higher value for the highest rotor speed generating steeper gradients can be
applied when using the IF optics.
9. If unsure about column lengths and rotor speeds, simulate SE data. To assist in selecting the appropriate configuration for the experiment, you can use the ‘Generate’ function of SEDFIT to assess the shape of the predicted SE profiles by simulating the approach to equilibrium at a given rotor speed. If this is based on realistic molar mass values and sedimentation coefficients of the free and complex species (for example, assuming equal concentrations of each), this will also indicate the expected minimal time to attain equilibrium.
A more advanced simulation with SEDPHAT is also possible, in which realistic levels of noise are added to theoretical equilibrium profiles calculated on the basis of hypothetical values for the binding constants. Re-analysis of this theoretical data set can be used to judge if the SE run will provide enough information to determine the correct binding constants with acceptable error estimates. The latter requires slightly more familiarity with the software than introduced in the present protocol, and the reader is referred to the extensive SEDPHAT help section at http://www.analyticalultracentrifugation.com/sedphat/sedphathelp.htm. In brief: (1) Use SEDFIT to generate a template data set for a non-interacting systems using the same rotor speeds and column lengths as those to be tested, with a dummy single sedimenting species model; (2) Make the xp-files and load the data in SEDPHAT as if doing an analysis of an interacting system; (3) choose the appropriate model and enter hypothetical concentrations and binding constants; (4) switch off any baseline parameters in the xp-files; (5) execute a RUN command; (6) Use the function "Save Fit Data" to save the theoretical curves. Finally, a similar procedure in SEDPHAT can be used to examine the kinetics of the approach to equilibrium for an interacting system with given kinetic rate constants of the interaction. This proceeds similarly as the SE simulation, but using a sedimentation velocity template for the approach to equilibrium.
1. Assemble the cell components according to manufacturer’s instructions. For the IF optics, sapphire windows are required, for the ABS system, use quartz windows. When assembling the components, determine if the window gaskets obscure the corners at the bottom of the sample and reference sectors. Frequently, some trimming with scissors is required to eliminate any shadow in the light path from the gaskets. This is very important in SE, because otherwise the region of the steepest gradients with the highest information content in SE cannot be observed. Select double-sector charcoal filled Epon centerpieces. (There are several alternatives: six-channel centerpieces permit more samples to be studied, but only at lower sample volume; for the IF optics, centerpieces with dual filling holes are available, or six-channel external loading centerpieces, which make thorough rinsing of the cell assembly easier after the ‘aging’ process (see next step)).
2a. For the IF optical detection only, mechanical stabilization of the cell assembly is required. Due to the exquisite sensitivity of the IF system to optical pathlength differences on the nanometer scale, rotor-speed and time-dependent signal offsets result from small deformation induced by the gravitational stresses. If these offsets are not removed or are unaccounted for in the analysis it may lead to erroneous parameter estimation.
This can be prevented by the following pre-conditioning procedure: a) Assemble the cells and torque to 120 – 140 inch´lb. Fill the cells with 200 microliters of water (the volume must be greater than the sample volume for the following SE experiment) and seal them. b) Insert the assemblies into the rotor, carefully align them, and centrifuge for 1 hour at 50,000 rpm. c) Stop the run and take the cell assemblies out of the rotor. d) Re-torque the cells, and observe if the screw rings have slightly loosened as a result of the centrifugation. e) Repeat steps b, c and d until no movement of the screw ring is required to reach the proper torque. f) Insert the cell assemblies in the rotor, insert the rotor in the centrifuge chamber, and install the optical arm. g) Accelerate the rotor to 50,000 rpm and configure the IF optical system. h) Set up ten alternating rounds of 2 hours centrifugation at 50,000 rpm and 1 hour centrifugation at 3,000 rpm. During each 50,000 rpm run, take a series of 10 IF scans in 5 minute intervals. This step can be conveniently set up as an ‘equilibrium method’ in the AUC operating software to run overnight. i) Establish that the scans in successive series show no change in RMS error, indicating that no further mechanical change in the cell assembly occurs. The newest version of SEDFIT allows users to determine whether successive scans overlay. This can be done by loading all the IF scans and selecting “Sort EQ data” in the Loading Options menu. Answer “no” to the next two windows (‘subtract blank scan?’ and ‘make xp files?’) and a plot of appears with Y= rms error and X=time. This completes the mechanical stabilization or ‘aging’ of the cells.
If the cells are to be stored before use, leave them filled with water. Important: do not disassemble the cells before their use. Only immediately before use, the remaining water can be aspirated by suction applied to small diameter tubing, and then filling the sample and reference solutions, as described in the following step.
2b. Acquire speed-dependent water blank scans for each cell. For the IF system only: Take ‘aged’ cells (still loaded with 200 ml water per sector), insert the rotor and mount the optical arm, and centrifuge at the first rotor speed selected for the SE experiment. Adjust the parameters for the IF optics in the ‘Laser setup’ for each cell (for criteria see Step III.8. of the SV protocol). After 1 hour of centrifugation, take a set of 10 IF scans (in the velocity mode with time interval 10 sec). Repeat at all other rotor speeds selected for the SE experiment. These scans will serve as ‘water blanks’.
Important: The settings of the IF optics, the particular rotor used, and the timing of the laser must remain unchanged for the remainder of the SE experiment for the water blanks to be valid. In order to reproduce these settings, make a note of each of the laser setup parameters for each cell. (This condition is certainly a weak point of using experimental water blanks, since removal and re-insertion of the cells into the rotor and the laser attachment into the rotor chamber may cause slight shifts. However, in particular when these steps are performed by the same operator, the reproducibility is usually sufficient.)
Stop the run, carefully remove the water from the cell assemblies through the filling holes, without causing any mechanical change in the assembly. For example, use suction applied through a small tube connected to a vacuum flask. Rinse both sectors of the cell with reference buffer and remove the buffer. This will prevent small changes in the buffer composition from residual water when filling the cell assembly. Then load with sample and reference buffer solutions.
This step II.2b is not essential in experiments with a sufficient range of rotor speeds and column length (the default recommendations) to permit computational determination of the radial-dependent baseline, in particular when IF and ABS detection are combined. However, the experimental determination can be both a safe-guard against unexpected profiles that do not meet the computational requirements, and a test for consistency of the data analysis.
1. Mix the samples in Eppendorf tubes. Fill the cell assemblies with the sample at the volumes planned in Steps I.5., I.6., and I.7. For the ABS system, fill the reference buffer to a volume 10 microliters larger than that in the sample sector. When using the IF system or a combination of ABS and IF, precisely match the reference buffer in volume (and chemical composition) to the sample. Seal the cell assemblies as described in Step III.3 of the SV protocol.
2. Insert the cell assemblies into the rotor, weigh and install the counterbalance, and align the cells at the scribe marks. Insert the rotor into the rotor chamber and mount the optical arm. (NOTE: when acquiring absorbance data below 400 nm, the high-pass filter lever should be parallel with monochromator housing.) Evacuate the chamber, and set the desired temperature on the centrifuge panel.
Determine whether or not the optical system is reliably radially calibrated. If not, select a rotor speed of 4,000 rpm, and perform the radial calibration of the required optical system. (Note that this can be done with the sample in place for a SE experiment, in contrast to the SV experiment.) A new radial calibration should be necessary only after the optical system has been changed or serviced, and with unchanged optical setup, the calibration should be valid for different rotors.
When the vacuum permits, accelerate the rotor to the first (lowest) rotor speed selected in Step I.8. Note that the equilibrium experiment does not require the temperature equilibration period that is essential for SV.
3. In exceptional cases when studying proteins of limited stability, an initial overspeeding period may be required to shorten the experimental time. For overspeeding, increase the rotor speed to threefold the first selected equilibrium speed, and set up a sequence of scans (e.g., absorbance at 280 nm) as a velocity method to monitor the concentration gradients. After a few hours, when the absorbance signal near the cell bottom has approximately doubled, stop the scanning and drop the rotor speed to the first equilibrium speed. (Any sedimentation creating excess concentration at the bottom of the solution column will significantly prolong the experimental time and may lead to irreversible or slowly reversible aggregate formation.)
4. Prepare the scan settings file on the AUC control computer. Specify the rotor type, set the centrifugation time to ‘hold’ mode, and set the run temperature. Set the parameters of each cell to the absorbance (and/or interference, respectively) equilibrium mode. Set the radius interval for scanning from 6.6 to 7.25 cm (for 180 microliter columns), and the wavelength to 280 nm.
5. In the ‘Options’ settings, select ‘Acquire intensity data instead of OD data’. This will separately store the radial-dependent light transmission through the reference and the sample sector, respectively, and provide a rational criterion for the appropriate data analysis range. The intensity scans (which will be saved in ‘*.ri*’ files) can be transformed to absorbance scans easily at any later time with SEDFIT without loss of information. In the same menu, set the number of overlays to 3.
6. For each cell, specify the scan ‘Details’: Enter as additional wavelengths 250 nm and 230 nm. Set the radial step size to 0.001 cm and request 20 replicates in the stepping mode. Specify if using the 2 (default), 6, or 8 channel centerpieces. Specify a new directory name for data storage. When using the IF system, set the inside and outside radius to encompass the complete solution column. Do not change the laser settings after water blanks have been acquired. Verify the ‘Laser Setup’ parameters are as noted in Step II.2b. Only if no water blanks were taken (for computational determination of the radial-dependent baseline), set up the laser timing and imaging parameters in the ‘Laser Setup’ (for criteria see III.8. of the SV protocol) and make a note of the settings for each cell.
7. Save the scan settings file and start a single scan to verify the quality of scans. When the ABS data acquisition is in intensity acquisition mode, as recommended above, two profiles will be shown for each cell: the radial-dependent reference transmission and the sample transmission. Both menisci should be visible; they can be identified as a local dip in the transmitted light. The scans should extend from the air-to-air region in front of the meniscus to the shadow region at the end of the solution column. The radial position r* near the bottom of the cell where the reference intensity starts to decline (from the shadow of components of the cell assembly) will mark the maximum radius that can be potentially considered for analysis. Write down this value for later use. The transmitted intensity through the sample, at least for one of the selected wavelengths, should be between 10% and 90% of the reference intensities. With time, the intensity transmitted through the sample close to the meniscus will increase, approaching that of the reference, and will strongly decrease or disappear close to the bottom, indicating the accumulation of the protein.
8. Set up and run a ‘Method’ for scanning. Described in the following are two alternative routines for setting up a method scan. The first routine involves manually stepping the instrument to the next highest speed once data for all cells in sedimentation equilibrium has been collected. The second routine allows for a scheduled automatic increase in rotor speed.
8a. Routine 1: In the ‘Methods’ menu for SE, start with the lowest rotor speed and set up a sequence of scans in six hour intervals for several days (‘delay condition’ is 6:00, ‘number of scans’ setting is 1, and enter the desired run temperature). Start a ‘Method’ scan. Allow scanning to continue at this first speed until the material in the cells reach sedimentation equilibrium.
To check for this condition in real-time, all scans collected by the absorbance optics or all scans collected by the interference optics (but not both types) can be loaded into SEDFIT. Under the “Options/ Loading Options” drop-down menu select “Test Approach to Equilibrium”, answer the resulting dialog boxes that appear accordingly (see SEDFIT help website). A plot will be generated which shows the root-mean-squared difference between each scan acquired for each cell and wavelength compared to the latest respective scan that was acquired. This plot will be automatically updated once new scans become available. The experiment should be considered to be at equilibrium once the rms difference asymptotically approaches a constant value. Further, this value should be at or below the level of noise in data acquisition (< 0.01).
A rigorous criterion can be formulated with the F-statistics, requiring that the rmsd difference between successive scans to be statistically indistinguishable. However, the visual inspection of the entire time-course of the approach to equilibrium, which should be a smooth asymptotic approach, is equally important. For an example, see http://www.analyticalultracentrifugation.com/eqtest.htm.
A frequent beginners mistake is to use too short time intervals for checking equilibrium (e.g., 3 hour intervals for long solution columns leaves too little time for sedimentation to proceed), and only to consider the difference between two successive scans. This can easily mislead to belief sedimentation equilibrium has been reached, when, in fact, it may still be significantly different from the true final distribution. It is important to note that concentration gradients that are still significantly away from equilibrium may already look quite exponential, and the deviation may not be caught in the data analysis and result in false parameter values. Therefore, getting a good fit in the data analysis cannot be taken as a confirmation of being sufficiently close to sedimentation equilibrium.
If the rms difference does not appear to have leveled off at a constant value, sedimentation equilibrium is not yet attained and centrifugation at the current rotor speed must be continued. Note that in this assessment the radial range considered for comparison will play a role, and should exclude the regions of optical artifacts and non-linear instrument response.
If equilibrium is not attained after what should have safely been sufficient time (e.g., two or three times the normal time required), consider whether proteolytic degradation or aggregation may take place, which will continuously change the distribution of soluble material. In this case, one may have no choice but to work with imperfectly attained equilibrium, accept the data and to proceed with the experiment, hoping that the sample degradation proceeds at a timescale much slower than sedimentation and diffusion of the sample, such that the loss of material is ‘adiabatically proceeding through mechanical sedimentation equilibria’ (– a case with the only disadvantage of not being able to use mass conservation constraints in the analysis). To confirm the presence of protein degradation, see Step III.10. Finally, it is advisable in this case to perform the interaction study by sedimentation velocity, which takes place on a much shorter time-scale and offers more tools to address the problem of slow protein degradation.
If equilibrium has been reached, stop the scanning and in the ‘Methods’ box, replace all previous rotor speeds, with the next higher speed. When using the IF optics, verify that the laser settings remain unchanged. Start a new set of scans and verify that this will accelerate the rotor. Repeat this step for remaining rotor speeds.
8b. Routine 2: As an alternative to 8a: For proteins that are known to be sufficiently stable, and in the absence of factors that could significantly extend the centrifugation time (e.g., high viscosity buffer, very large protein complexes > 200 kDa, very elongated protein shape leading to an unusually small diffusion coefficient, or working with diluted stock mixtures of reactants that exhibit very low chemical off-rate constants), the ‘Methods’ for the SE can be configured in one step to go through a sequence of rotor speeds. For 170 microliter columns, start with the lowest rotor speed and request 8 scans in 6-hour intervals. Continue with the next higher rotor speeds, for each requesting 6 – 8 scans in 6-hour intervals. (Frequently the equilibrium at the second and third rotor speed is attained faster than the first.) At the end of the highest rotor speed, append a line with the delay condition ‘hold’. Start the ‘methods scan’. This will automatically collect the necessary set of scans and switch the rotor speeds during the next several days.
Attainment of equilibrium can be observed during the run with SEDFIT as described above, and/or verified after the run is finished (or better while the highest rotor speed is still on hold). The obvious disadvantage of this routine is that it does not offer the possibility of extending the sedimentation time retroactively, except for the highest rotor speed. However, if, during the run, it is found that equilibrium is not going to be properly attained (for example, if from the time-course of rmsd difference to the last scan it can be discerned that not sufficient time is available for the asymptotic approach, or the rmsd differences are still too high), this scan ‘Method’ should be stopped, and restarted with a modified protocol that allows for sufficient scans. (In this case, account for the fact that a number of scans have already been taken.)
9. Stop the run, once equilibrium profiles at all rotor speeds have been acquired.
10. Check the stability of the protein. It is generally advisable to recover the sample for analysis by SDS-PAGE or mass spectrometry. This can show if proteolytic degradation has occurred. Alternatively, the solution can be re-suspended by carefully shaking the cell, and a short-column SV experiment can be conducted with the c(s) analysis indicating the solution state of the protein mixtures. (The latter is not done routinely).
At this point, if further study of the sample by dynamic light scattering is desired, the centrifugal cell can be carefully (without shaking too much) removed from the rotor, opened, and a small (20 – 30 microliters) aliquot can be drawn from the top of the solution column just underneath the meniscus. This can take place such that only laminar movement of the liquid occurs inside the centerpiece, leaving much or most of the concentration gradient intact. It should be noted that this procedure will produce a sample biased towards the smaller species, but it is usually of excellent quality with regard to the removal of aggregates and larger particles that interfere with light scattering.
11. When using the IF optics, take another water blank: Without disassembly, carefully remove the sample and reference solutions, rinse the cell assembly with water, insert water, seal, and re-centrifuge and scan the water-filled cells at the rotor speeds of the equilibrium experiment. This will generate a new set of water blank scans to be compared with the initial water blanks to verify the optical stability of the cells and the acquisition system. This step is not required if the computational determination of the radial-dependent baseline is chosen.
12. Clean the cell assemblies as outlined in Step IV.2 and IV.3 of the SV Protocol, except when using ‘aged’ assemblies. The cleaning of these can take place without disassembly by exhaustive rinsing. This is facilitated with six-channel external loading cells and the dual filling-hole double-sector cells. This allows the pre-conditioned ‘aged’ cells to be re-used without new ‘aging’ cycles, as long as they remain mechanically unaltered. Store the ‘aged’ cells filled with water.
When executing this data analysis for the first time, visit the website http://www.analyticalultracentrifugation.com/sedphat/sedphat.htm and familiarize yourself with the help system of SEDPHAT, the introduction to the organizational structures and the screenshots for the specific models. It can be useful to have access to the online help system while going through the following analysis steps.
Note that there is additional tutorial material available online at the SEDPHAT website at http://www.analyticalultracentrifugation.com/sedphat/sedimentation_equilibrium_for_interacting_systems.htm. In particular, this includes the step-by-step tutorial for the study of a protein-DNA interaction at http://www.analyticalultracentrifugation.com/sedphat/DNATutorial_2.htm. Novices may want to consider downloading the example data and following simultaneously the following explanations and the notes and screenshots at this website.
Preparing the data for analysis.
To prepare the data for analysis in SEDPHAT, they must first be sorted into “experiment” (*.xp) files. This step takes place in SEDFIT, using the “Sort EQ data to Disk” function in the “Loading Options” menu. An xp file is usually comprised of the equilibrium scans for a single cell at one wavelength. It may contain only data from a single rotor speed, or contain data from all three rotor speeds (the latter termed ‘multi-speed equilibrium experiment’). The xp files that are created in this manner (described in the following) can then be brought into SEDPHAT individually or collectively for “Local” or “Global” fitting.
1. 1a. Collect the data. Copy the data folder of the entire run, back it up, and copy the data onto a computer dedicated for the data analysis.
The location of the relevant data depends on which routine in Step III.8 was used for data collection, i.e. whether routine 1 was used with the start of several ‘Methods’ or routine 2 with all data acquisition combined into one ‘Method’. Each time a set of scans is started, the XLA/I program creates a new subdirectory with the current date and time and stores all files numbered sequentially, with different extensions for each cell. Unfortunately this makes it difficult to determine easily the wavelength and rotor speed for each scan, and which ones were the latest ones at any given rotor speed, i.e. closest to equilibrium. However, this task can be accomplished with a SEDFIT function described in Step IV.1b.
If multiple data directories were created (i.e. multiple ‘Methods’ were used), it can be convenient to merge all data files into one folder before proceeding. In order to avoid overwriting files that have the same filename, there are two options: (1) Load the data from each folder into SEDFIT (ignore the warning about the temperature fluctuation that may occur), and use the function “Save Raw Data Set” from the OptionsàLoading Options menu. Accept the “save data with wavelength/speed information in filename …” option. This will change the filenames and can allow files from different folders to be merged into a single folder eliminating conflicting filenames. However, if different methods were started containing data at the same rotor speed, conflicts will still arise. In this case, use the following option: (2) Using the same function, do not accept the option “save data with wavelength/speed information in filename …”. This will, instead, use the original filename but with a preceding ‘x’. These data can then be copied into the folder containing all other scans. If needed, this procedure can be repeated creating multiple preceding ‘x’ characters until no conflicting filenames exist.
Note that 6-channel centerpieces require a slightly different procedure as described below in Step IV.1e.
1b. Process the ABS scans into xp files. Load all the ABS scans from all the cells (all speeds and wavelengths) into SEDFIT (DataàLoad New Data). This may be *.ra* (absorbance) or *.ri* (intensity) files. Then, from Loading Options menu of the Options tab, select “Sort EQ Data to Disk” and follow the SEDFIT screen prompts to generate xp files which the analysis software SEDPHAT will recognize. This process is described in detail in the Appendix “Sort EQ Data to Disk” below. SEDFIT will again allow you to examine the scans in equilibrium and this time will give you the choice of whether to create and include specific scans in a multi-speed experiment file with a *.xp extension This step will also allow you to convert intensity scans into absorbance scans. Choose not to “subtract blank file”, but to “create xp files”.
1c. Process the IF scans without water blanks (using calculated TI noise as computational blank). If interference data have been collected at several rotor speeds creating different curvatures as outlined above, and/or if combined with absorbance data in a global analysis, then cumulatively there is usually enough information to computationally calculate the radially-dependent baseline (see Vistica et al.). This option is selected when fitting for TI-noise (see below). In this case, it is unnecessary to subtract experimental water blanks.
To sort the IF data collected on your samples, load all the scans from all cells and rotor speeds into SEDFIT. Choose “Sort EQ data to Disk” from under the Loading Options menu as in Step IV.1b. Choose to “make xp files” and not to “subtract blank scan” from the appropriate resulting dialog boxes.
1d. Process IF scans for blank subtraction. If the radial-dependent baseline noise for your sample scan is to be corrected for by subtraction of a water blank, then it will be necessary to process your blank scans and sample scans separately, and then perform the subtraction in a separate step. Water blanks are essential when data from only a single rotor speed are available. They also may be used in addition to the computational blank correction.
In this case the calculated TI noise profile should only lead to a refinement of the fit by accounting for systematic high spatial frequency components (e.g., blips), but should not exhibit significant slopes or curvature.
Pre-process blank scans: Load IF scans of water blanks collected for a single cell and rotor speed in SEDFIT. Select upper and lower fitting limits. The limits should be set such that they cover the entire radial range in which your analysis of protein samples will be executed (keeping away from the artifacts near the cell bottom and meniscus). Select the “Non-interacting Discrete Species” model and choose to fit only for the baseline, RI and TI noise in the parameters box (deselect any components, and meniscus or bottom position). Perform a Run to calculate the average trace, which will be contained in the TI noise. Notice the rmsd should be at a value well below 0.01. Save the TI Noise (in the SEDFIT menu DataàSave Systematic Noiseàsave Noise in File) into a labeled folder. Repeat this process for each cell and rotor speed.
In analogous manner, the water blanks before and after the experiment can be loaded and compared, and an average may be used for correction of the equilibrium scans.
Pre-process sample scans: Load all the sample IF data collected for all sample cells at all speeds into SEDFIT and sort them as in Step IV.1c above except opting to not “make xp files”. This process will label and save the last scan at each speed into a designated directory.
Subtract blank scan from sample scan: In SEDFIT, load a sorted interference scan from this designated directly (i.e. a single cell at a single rotor speed, determined in the previous step to be the equilibrium scan) by using the SEDFIT function “Load Sedimentation Equilibrium Data”. Subtract the respective water blank scan (created as described in the preceding paragraphs), with the function Optionsà Loading OptionsàSubtract Blank Scan. Save the blank-subtracted scan. Repeat this for each IF data set.
Finally, create multi-speed xp files in SEDPHAT. Open SEDPHAT, choose “Load Multi-Speed Equilibrium Data” from the Data drop-down menu and then simultaneously select the three sorted, blank-subtracted IF files corresponding to a single cell at all three rotor speeds. This multi-speed equilibrium file is saved as an experiment with an *.xp extension. This format will allow the user to later load this file (in multiple ways including drag-and-drop and point-and-click) for an individual analysis or as part of a global analysis.
Alternatively, one may choose to subtract a single blank scan from all sample data for a particular cell obtained at one or more rotor speeds. To do this, load all water blank IF scans (irrespective of cell number or rotor speed). Sort them as in IV.1b. Load all sample data scans for a particular cell (multiple speeds) and sort them as previously stated, but select to “subtract blank file”. Indicate which file should be subtracted from the sample scans and save the blank-subtracted file into an indicated directory location.
1e. When using 6-channel centerpieces, open all scans (absorbance or interference, but not both at the same time) into SEDFIT. From the “Loading Options” menu under the “Options” tab, select “Save 6-channel Raw Data in 3 Subsets”. Then Load all scans (multiple speeds and signal) within a subset (inner, middle, outer sector) into SEDFIT (version 10.11 or newer) and “Sort EQ Data to Disk” creating multi-speed equilibrium files. For water blank subtraction, follow the Step IV.1d above for each of the separate subset files.
Analyzing the sedimentation profiles of the individual components to determine their extinction coefficients and apparent molar mass M*
2.
Open SEDPHAT and assemble all the xp files associated with species
“A” only. For example, load the xp file with the equilibrium scans
at 280 nm (which will include scans from all 3 rotor speeds) of the cell with
the sample containing only component ‘A’. Graphically estimate the meniscus and
bottom. For the left fitting limit, exclude the artifacts close to the
meniscus. For ABS data, determine the right fitting limit as the highest radius
where the maximum absorbance is < 1.5 OD, but not exceeding the radius r*
determined in
Step III.7. Also, avoid regions of very steep gradients. For IF data, set
the right fitting limit to exclude regions of optical artifacts near the bottom
of the cell. In the experiment (‘xp’) parameters, mark the bottom to be a
floating parameter. Specify the known extinction coefficient at 280 nm and the
buffer density. Enter the ![]() of the component (or an estimate), or an operationally defined
of the component (or an estimate), or an operationally defined ![]() ; set this value both for the local parameters in the ‘xp’
parameter box, as well as for the global parameters in the OptionsàSet
vbar*rho menu.
; set this value both for the local parameters in the ‘xp’
parameter box, as well as for the global parameters in the OptionsàSet
vbar*rho menu.
Load the additional xp files from the other two wavelengths and the IF scans and repeat this process.
The extinction coefficients of the other wavelengths may not be known. In the xp parameter boxes of these data, enter an initial estimate of half the 280 nm extinction coefficient for the 250 nm data, and fivefold the 280 nm extinction coefficient for the 230 nm data, and mark this parameter to be floated in the non-linear regression.
For the IF data, use an equivalent extinction coefficient of 2.75 times the molecular weight (e.g., 275000 for a 100 kDa protein) as a starting guess. For non-glycosylated proteins, this value can also be used as a fixed, known parameter in conjunction with a floating extinction coefficient at 280 nm.
Since all data are from the same cell, there is only one unique meniscus and bottom location. This should be reflected in setting up links between the floating bottom position of all ABS data sets. Note, in a global analysis like this the first xp file that you load is considered experiment 1 (in the software terminology), likewise, the second xp file loaded is considered experiment 2, etc. Accordingly, redirect the floating bottom parameters of xp #2 and #3 (the 250 nm and 230 nm data, respectively) to the xp #1 (the 280 nm data). Linking the bottom position of the IF data set to that of the ABS data sets is usually not recommended since the radial calibration of the different optical systems is usually imperfect, causing negligible errors in the normal analysis but potentially larger errors if these parameters are constrained to be exactly the same.
Finally, save the configuration by selecting from the data drop down menu “copy all data and save as new configuration”. Similarly, assemble a like configuration for component “B”.
3. Configure the analysis: Read the configuration for component A into SEDPHAT. Make sure all xp-files of all signals are loaded. Select the model “A (single species of an interacting system)”. In the Global Parameters: Enter an estimated molar mass, and mark it to be floated in the fit. Switch on ‘mass conservation’, and ‘vary local concentration’. Uncheck the sedimentation coefficient (which is not meaningful for the equilibrium experiment). Set the concentrations in the Local Parameters: For the experiment at 280 nm (or that with known extinction coefficient, respectively), link the concentration to itself, and enter the loading concentration in micromolar units. For all other experiments, link the concentrations to the first experiment with known extinction coefficients. This redirection will ensure that only one unambiguous value for the concentration for each cell is used, which will permit calculation of the extinction coefficients. Update the configuration.
4. Fitting: First, test the starting estimates of all parameters with the ‘Run’ command. If necessary, change some estimates to ensure that the theoretical curves at this stage are already in the same order of magnitude as the experimental data. Fit the model. Repeat the fit with alternating optimization methods (Simplex and Marquardt-Levenberg). After the non-linear regression has converged, update the configuration, and document the fit by copying the SEDPHAT window display.

Assess the quality of the fit, using the local rms deviations as a measure (they
should be < 0.01, within the noise of the data acquisition), and the assessment
how systematic the residuals are. For example, in the following plots the
residuals are borderline acceptable in the top, unacceptable in the middle, and
excellent in the bottom panel.
However, the total loading signal also has to be taken into account: If, for example, the concentration gradient amounts to several fringes in the IF signal from meniscus to bottom, then residuals larger than 0.01 fringes can indicate a very good fit. Also, when using water blanks, allowance has to be made for imperfections in the reproducibility of the blank, which may be judged from comparison of the ‘before blanks’ and ‘after blanks’.
For assessing the quality of fit when using TI noise as a computational blank of the IF data, an additional important consideration is the shape of the calculated TI noise (as indicated in the black line drawn in the plot of IF data [note that it will have an average of zero, which may require manual adjustment of the vertical plotting range to make the solid black line visible]). It should consist of a more or less horizontal line which will have blips and bumps, but be overall flat and not exhibit significant overall slopes or curvatures, in particular not towards the right fitting limit. The latter would indicate that TI noise parameters are compensating for erroneous Mw parameters (or erroneous model).
5a. If the fit is satisfactory,
note the best-fit apparent molar mass value, M*A . If the
molar mass scale was based on an operational ![]() , transform the molar mass to the ‘true’ molar mass MA
on the ‘true’
, transform the molar mass to the ‘true’ molar mass MA
on the ‘true’ ![]() scale with
scale with ![]()
![]() . The determined molar mass MA should be
within a few percent of the molar mass from sequence or mass spectrometry, with
deviations usually originating from errors in the estimated
. The determined molar mass MA should be
within a few percent of the molar mass from sequence or mass spectrometry, with
deviations usually originating from errors in the estimated ![]() . Also note the best-fit values of the extinction
coefficients. Along with M*A , these will be used in the
subsequent interaction analysis.
. Also note the best-fit values of the extinction
coefficients. Along with M*A , these will be used in the
subsequent interaction analysis.
5b. If the fit is not satisfactory, some of the most common problems are: 1) The amplitudes of the fitted distributions are very far from the measured profiles. If so, the loading concentrations were initialized poorly. 2) Sometimes the ABS baseline offsets are not constant and shift with rotor speed. In this case consider the ‘fit RI noise’ option to permit individual baselines. 3) Mass conservation is not strictly fulfilled. If so, make sure the bottom position was an adjustable parameter during the fit, and if necessary remove the mass conservation constraint. When fitting without mass conservation constraint, uncheck the bottom position to make this a fixed parameter for all xp’s. 4) The component is self-associating. In this case, switch to a self-association model and try to get a better fit. For example, use the monomer-dimer model and initialize the log(Ka) to the negative log10 of the loading concentration (e.g., leading to a value of 6 for micromolar, or 5 for 10 micromolar loading concentration). Note, moderate or strong self-association should correlate with M*A significantly higher than the true molar mass of this species, while weak self-association may not directly apparent this way. When analyzing the data with self-association models, it may be necessary to fix M*A to the theoretically expected value, in order to avoid strong correlation of the Mw and log(Ka) parameters. In case of self-association, the experiment should be repeated with several loading concentrations of this component. Also, self-association, if unexpected, should be reconciled with separate sedimentation velocity experiments, where it should show in the form of a concentration-dependent weight-average s-value. This will help to discriminate the self-association model, and to distinguish self-association from the presence of impurities. 5) Impurities of small molar mass or aggregates are present; inspect SDS-PAGE, HPLC elution profiles, or c(s) SV distributions of the recovered sample. Sometimes, impurities will exhibit significantly different signals at particular wavelengths: for example, contaminations with peptides containing no aromatic amino acids will show strongly at 230 nm, not at all at 280 nm, and very little in IF – in this case, it can be advantageous to exclude the 230 nm data from consideration. 6) Thermodynamic non-ideality is affecting the sedimentation at high concentration; this will generate a distinctly lower slope of the profiles than predicted by the fitted lines at higher concentrations.
6.
Repeat Steps IV.2 – IV.5 with the second component.
Use the same ![]() as an operational quantity, and derive the apparent molar
mass M*B of the second component based on the same
as an operational quantity, and derive the apparent molar
mass M*B of the second component based on the same ![]() scale as with the first component. In this way, although the
apparent molar mass values appear different than the known sequence molar mass,
they are valid for the interpretation of the centrifugation experiment on the
particular
scale as with the first component. In this way, although the
apparent molar mass values appear different than the known sequence molar mass,
they are valid for the interpretation of the centrifugation experiment on the
particular ![]() scale, and they are directly additive in complex formation,
for example, M*AB = M*A+M*B.
scale, and they are directly additive in complex formation,
for example, M*AB = M*A+M*B.
Determining the equilibrium binding constant of the hetero-association
7.
Load the data. For each cell with mixtures, assemble
the scans at each wavelength (and IF) into SEDPHAT configurations as described
above in
Step IV.2. Set the meniscus, bottom and fitting limits as before in
Step IV.2. Mark the bottom position to be a floating parameter, and link
different xp’s from the same cell but different ABS wavelength to have the same
meniscus and bottom. Use the same operational ![]() and enter the predetermined extinction coefficients for the
respective wavelength (unmarked as fixed parameters). Save the Configuration
(use the ‘copy all data and save as new config’ function).
and enter the predetermined extinction coefficients for the
respective wavelength (unmarked as fixed parameters). Save the Configuration
(use the ‘copy all data and save as new config’ function).
8. Set up the parameters. Select the ‘A + B ßàAB Hetero-Association’ model. Load xp files of all mixtures in SEDPHAT. Not all wavelengths are absolutely required, but the loaded set should contain the most informative signals (usually 230 nm for the lower concentrations, 250 nm for the highest concentrations). If possible, IF data should be combined with at least one ABS signal from the same cell.
In the global experiment parameters, enter the predetermined values of the apparent molar masses for each component, M*A and M*B. Mark the ‘atot’ and ‘btot’ field to optimize the local concentrations in each cell. Switch the mass conservation model on. Set the log(Ka) field to an expected value (for example, – 5 for binding constant of KD = 10 micromolar). Uncheck the s-values for all parameters. Set the incompetent fractions to zero in the ‘incfA’ and ‘incfB’ fields, and uncheck these fields. Uncheck ‘add non-participating species’. Check ‘Mass Conservation’, if mass conservation constraint could be applied for both analyses of the individual components A and B. (But even if the mass conservation is not fulfilled for A and/or B, it is advisable to use mass conservation as an approximation in a first stage of the fit, i.e. to get the concentration and other parameters in a good range, and then in a second stage of the fit to eliminate the mass conservation constraint.)
Dependent on the choice of the titration or dilution series for the mixtures in Step I.5, in the global parameters either toggle to the ‘both A,B in micromolar concentration’ field, or to the ‘A in micromolar conc and B/A molar ratio’ field. The former is advantageous for titration series, because it allows to use the constraint of a constant concentration in A (or B) among the data of different cells, while the latter is advantageous for dilution series, because it allows to constrain the molar concentration ratio to remain constant among different cells of this dilution series. (Even though it is very difficult to know exactly the molar ratio, it is certain that the molar ratio remains constant in a dilution series.) These constrains are extremely useful to eliminate the correlation between the estimate of the binding constant and the ratio of the loading concentrations, which exists for interactions between proteins which are too similar (M*B < 2´M*A or M*A < 2´M*B) or too dissimilar (M*B > 10´M*A or M*A > 10´M*B) in size.
In the local parameters, for each cell enter the loading concentrations of both components (in micromolar units), or component A (in micromolar units) and the ratio B/A (molar ratio), respectively. In doing so, establish links redirecting the loading concentrations for data sets of different signals from the same cell to point to only one ‘experiment’ (or ‘xp’ data channel) for each cell, and establish links that reflect the titration or dilution configuration among the cells (for example, in a titration series with constant A by redirecting the concentration parameter of A of all cells to that of xp #1) .
Update the configuration (including xp’s).
9. Fitting. Use the ‘Run’ command to verify that the initial estimates of the parameter values produce theoretical distributions in the correct order of magnitude. If the solid lines indicating the theoretical model do not at least appear in the same plot following the same general trends as the data, double-check that the correct extinction coefficients are used in each xp, and manually explore different concentration values. When it is difficult to get reasonable starting conditions, sometimes the following strategy helps: Set the bottom values for all cells to a graphically determined value (or 7.2 cm for double-sector cells), and uncheck this parameter in all xp parameter boxes. Also, uncheck the log(Ka) in the global parameter box, such that the concentration values are the only unknowns to be fitted. For samples with equimolar loading concentration, set the B/A molar ratio to 1 and fix this value. Then use the Fit command to ‘pre-optimize’ the concentrations and get them in the right ballpark. After that, revert back to the floating log(Ka) and floating bottom parameters. Also, if applicable, revert back to a floating B/A molar ratio (since the precise molar ratio usually cannot be known accurately enough a priori to serve as a fix constraint).
Fit the model (execute the fit command) with alternating optimization methods (to be switched in the OptionsàFitting Options menu). The simulated annealing method is particularly powerful to find global minima, since the error surfaces of sedimentation equilibrium models for interacting systems typically exhibit shallow local minima that can be difficult to deal with. After parameter values have converged, and no improvement can be found with either optimization method, save (update) the configuration and document the result. Assess the quality of the fit, using the criteria outlined above in Step IV.4.
If the fit is not acceptable, there are several possible explanations: (1) A possible consideration is often the existence of a subpopulation of material incompetent to associate. (This can be tested by including ‘incfA’ and/or ‘incfB’ as floating global parameters. But critically inspect the returned values after fitting: A value of 0.5, for example, means that 50% of material was binding incompetent – a number that may be ruled out based on other knowledge on the protein, such as mobility shift or gel filtration data. In contrast, a value of 0.05 would indicate 5% incompetent material, which is hard to rule out by any method.). (2) In rare cases, mass conservation may not be fulfilled for the mixtures, even though it is fulfilled for each individual component. To test this, switch off mass conservation (see the additional comments in Step IV.5.b). (3) The sample may be contaminated with a contaminating species not participating in the binding reaction. This can be assessed from accompanying SV experiments, where c(s) can provide Mw estimates for this component, if the mass is outside the range of those of the individual components and the complexes. This species can be accounted for in the SE data analysis here using the ‘add non-participating species’ option. Note that the loading signal (directly in signal units) for this species in each xp will need to be initialized, and should be floated. (4) Alternatively, different association models may be explored. For example, if the theoretical lines are systematically not steep enough compared to the data in the region close to the right fitting limit, and if this mismatch increases with higher loading concentrations, an association model with larger complex is indicated.
10. Inspect the reliability of the fit in three ways: (1) If TI noise was used to calculate a baseline blank in either IF or ABS data, verify that it does not show significant overall slopes and curvatures towards the right fitting limit. If this is the case, there is a correlation between fitting the concentration gradients and the baseline, and the TI noise calculated blank should be switched off. (2) Visualize the contributions of the complex to the signal. This will reveal whether for the given experimental concentration range and the calculated KD any significant complex formation is predicted: If not, then the KD value certainly cannot be determined from the given data, and only a lower limit can be calculated. Vice versa, visualize the contributions of the free components – if they are essentially zero, i.e. if any one is not predicted to contribute significantly to the signal, then binding cannot be distinguished from stoichiometric binding and only an upper limit for the KD can be extracted from the data. The respective traces for these species can be switched on in the Display menu.
11. Perform a statistical error analysis on the determined binding constant. Different methods are available in SEDPHAT, but the most reliable and most rigorous approach is the projections of the error surface method (Bevington and Robinson 1992). (For ‘simple’ fits, the Monte-Carlo approach works well.) Although the absolute value of c2 (‘global red. chi-square’ in SEDPHAT) should not be interpreted without absolute knowledge on the noise in the data, the relative increase of c2 comparing the original and the new fit is a statistical quantity that can be evaluated using the F-statistics. Calculate the critical value for c2 (with the SEDPHAT statistics function). Set the value of the binding constant at a non-optimal value close to the best-fit parameter (for example, increase log(KA) by 0.05), fix the binding constant but float all other unknown parameters in a new fit. If the original fit was the best fit, then the current fit will have a higher c2. If the new value exceeds the critical value, then the modified binding constant is outside of the error interval. If it is less than the critical value, fix log(KA) at a value a little bit further from the best-fit value and re-fit. Repeat this until the critical c2 is exceeded, which indicates that one of the limits of the log(KA) error interval has been found. Reload the best-fit configuration and repeat the procedure by changing log(KA) in the other direction. An illustration of this approach can be found in (Schuck et al, 1999).
![]()
Appendix: Test approach to equilibrium
(SEDFIT version 10.11, April 2007 or newer).
1. Load all scans collected thus far (by either absorbance optics or interference detection, not both) in a new SEDFIT window.
2. Set upper and lower “fitting” limits. The lower limit should be to the right of the meniscus and the bottom fitting limit should be located at a position where the signal is linearly dependent on concentration. Optical artifacts must be excluded from the range between the fitting limits.
3. Reveal the drop-down menu under the loading options item of the Options tab and select “test for approach to equilibrium”.
4. Indicate whether the indicated scan (cell x, speed j) should be included in the plot of the ‘time-course to equilibrium’. This scan could be omitted for example if it appears that it will not be useful for data analysis perhaps because the signal is too low or there is insufficient curvature.
5. “Accept radial limits for comparison…” answer ‘Yes’ to this question if the limits (indicated by the red line overlaying raw data) are acceptable. Answer ‘No’ if the limits need to be adjusted.
6. Continue in this manner for the remaining scans at available rotor speeds.
7. “Real time update?” Answer ‘Yes’ if you would like the software to automatically update the ‘time-course to equilibrium’ plot with new scans that acquired at later times and rotor speeds. The user may choose whether to use the default limits already set previously for the additional scans loaded in the real-time update. The user may also choose whether to include scans that are acquired later for cells that had not yet been included in the time-course plot.
![]()
(SEDFIT version 10.11, April 2007 or newer). For screenshots of this function, see http://www.analyticalultracentrifugation.com/eqtest.htm.
1. Load all scans (from all cells, rotor speeds, and wavelengths) collected by either interference or absorbance detection (but not both types at once).
2. With the left mouse button, double-click (and then hit ENTER) to set up initial positions for the upper and lower radial fitting limits. The fitting limits should span the entire range of radii that will likely be included for data analysis (linear concentration range) and excluding the regions containing optical artifacts near the meniscus and cell bottom. They will be refined later.
3. From the “Loading Options” menu under the “Options” tab, select “Sort Equilibrium Data to Disk”, point to where the data should be saved (it is recommend that the data be saved in a folder entitled “SortedEQData”). Choose not to “Subtract Blank File” (as this option only allows to select a single blank file, which is not sufficient when handling data from different cells and/or rotor speeds). Accept the option to “make XP Files” – this will result in the creation of a multi-speed equilibrium file comprised of the final scan at each rotor speed for a particular signal. Input buffer density, protein partial specific volume and molar extinction coefficients or accept default values (these can be changed later) into the resulting dialog boxes. A plot will appear that shows the root-mean-square difference in signal between each scan and the latest respective scan acquired and a dialog box inquiring whether the sample had reached sedimentation equilibrium. For this judgment, see also the comments in Step III.8 above. Selection of “yes” will generate a data file comprised of the last scan at the indicated speed and wavelength. A new dialog box will appear inquiring whether to include that data set in a multi-speed equilibrium xp file.
4. Continue responding to the remaining dialog boxes indicating whether a particular cell attained sedimentation equilibrium at each indicated rotor speed and input signal, and whether to include the final scan in a multi-speed equilibrium xp-file.
![]()
References:
J. Vistica, J. Dam, A. Balbo, E. Yikilmaz, R.A. Mariuzza, T.A. Rouault, P. Schuck (2004) Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Analytical Biochemistry 326:234-256
P. Schuck and D. Millar (1998) Rapid determination of molar mass in modified Archibald experiments using direct fitting of the Lamm equation. Anal. Biochem. 259:48-53
P.R. Bevington an D.K. Robinson. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill, New York, 1992
P. Schuck, C.G. Radu, and E.S. Ward (1999) Sedimentation equilibrium analysis of recombinant mouse FcRn with murine IgG1 and Fc fragment. Mol. Immunol. 36:1117-1125