Example 1: Isopycnic density gradient sedimentation of a protein in 2.5 M CsCl
Example 2: Protein sedimentation in lower co-solute concentrations (1 M CsCl)
Example 3: Experimental data of a protein in 1.4 M CsCl
Example 4: Analytical zone centrifugation with isopycnic conditions
Sedimentation experiments are rarely performed in pure two-component systems with a macromolecule and a pure solvent. This has many reasons, one of which when dealing with proteins is commonly the need to stabilize the pH and add salts to screen long-range electrostatic forces. Normally, at low concentrations of small co-solutes such as the buffer components, this does not pose any further complication, as long as we take into account the effect of the co-solutes on the density and viscosity of the solvent.
In rare cases of very high co-solute concentrations, we may have to consider preferential hydration issues, and correct the buoyancy factor (either in form of an 'apparent' partial-specific volume (1-F'r) or in form of a density increment dr/dc). Usually, for example with proteins in PBS, these effects are negligible.
Sometimes, however, the sedimentation of the co-solutes themselves creates a dynamically evolving co-solute gradient and, as a consequence, a density gradient. Exploiting this effect has a long history. One such case is traditional isopycnic density gradient sedimentation, where a high concentration of, for example, CsCl has been used to create a self-forming density gradient that includes conditions of neutral buoyancy for macromolecules like nucleic acids (1), protein, protein-lipid complexes, carbohydrates, viral particles, etc. Another example is analytical zone centrifugation (band centrifugation), where a dynamic density gradient is formed by layering a lamella of lower density solution (containing the macromolecule) on top of a higher density solution.
A self-forming density gradient may also be created inadvertently, for example, when sedimenting proteins at high rotor speed in the presence of glycerol, sucrose, or high salt concentrations.
The significance of redistributing buffer components can be assessed by looking at the interference optical fringe displacement data when the reference solution is replaced by water.
It can be of interest to analyze what happens with the macromolecular sedimentation in the presence of such dynamic density gradients. This is of long history in the theory of sedimentation (6,7,8,9,10). The coupled co-solute and macromolecular sedimentation process has been included in Sedfit for simulating and modeling of experimental data. Details of the approach are published in (2) and practical aspects of these analysis in Sedfit can be found here.
To illustrate some basic features, let us look at three examples.
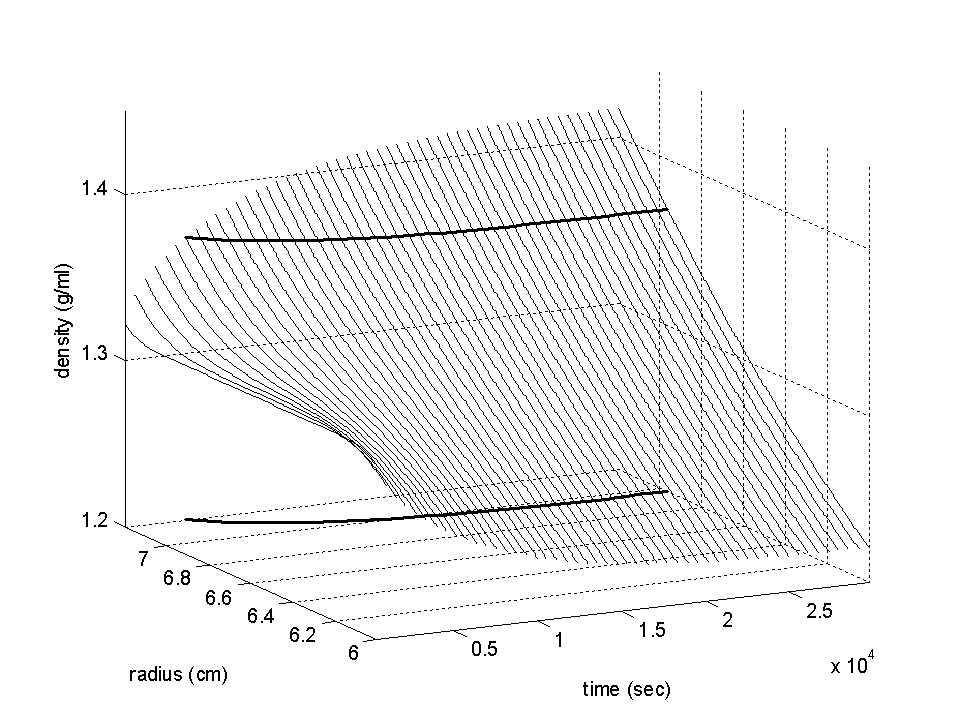
Example 1: Isopycnic density gradient sedimentation of a protein in 2.5 M CsCl
The first example is the simulated sedimentation of a protein of 500 kDa in a buffer with 2.5 M CsCl at 60,000 rpm. The density of a 2.5 M CsCl solution is 1.31 g/ml. However, the salt will redistribute with time and cause regions of lower density close to the meniscus, and regions of higher density closer to the bottom. The following graph shows the evolution of the density gradient with time.

Initially homogeneous, the distribution will approach a steady-state gradient after longer time. Indicated in the graph as a bold lines is the density where a protein would float (it is shown as intersection with the density data r(r,t) and again in a projection to the radius-time plane). An interesting feature is that the neutral buoyancy condition does initially not exist anywhere in the solution, which means that sedimentation of protein takes place everywhere, but that the neutral buoyancy point slowly peels of from the bottom of the cell, causing proteins to concentrate around this point (they will sediment or float, respectively, towards this point).
The protein distribution at different times is shown in the next plot:
You can see it starts initially like ordinary sedimentation, but then slowly
moves back out. After long times, the protein distributes like a Gaussian,
with a width determined by the molar mass.
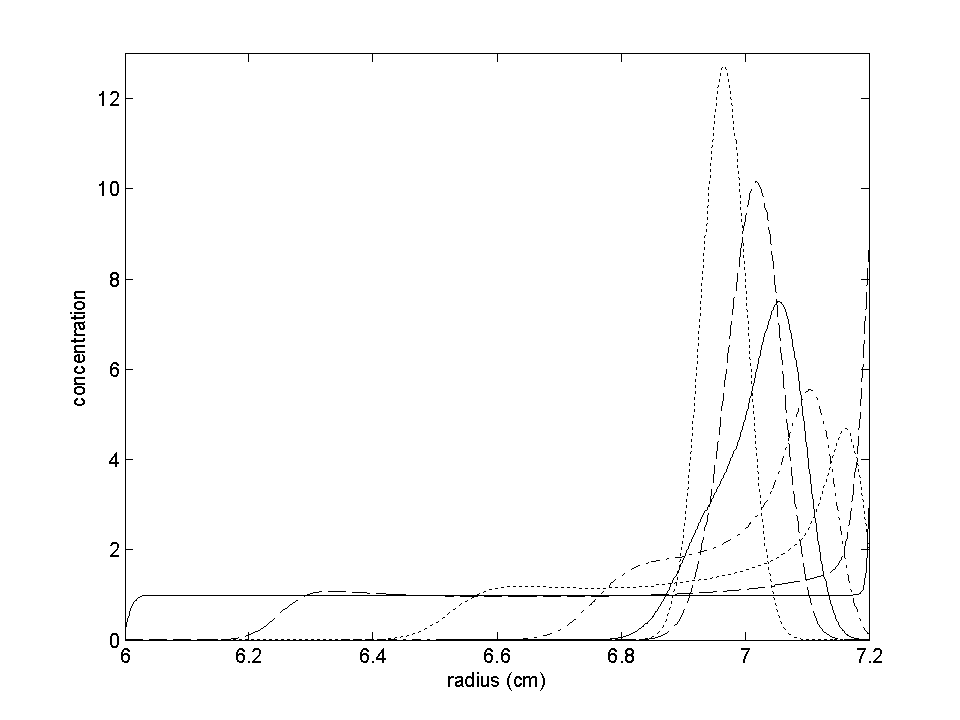
(For details, see ref 2). An interesting detail is that the 'plateau' regions can have two maxima - the main peak moving towards the neutral buoyancy point, but there's a second small peak just after the boundary (see below).
Example 2: Protein sedimentation in lower co-solute concentrations (1 M CsCl)
We don't need to go to extreme conditions of neutral buoyancy in order to observe significant effects of the co-solute redistribution on the macromolecular sedimentation. The second example is for a 200 kDa protein in 1 M CsCl at 60,000 rpm. The initial density is 1.126 g/ml, and neutral buoyancy will never be reached.
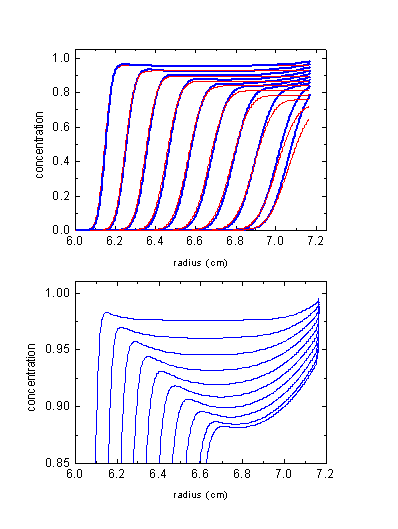
The blue lines show the calculated protein distributions, which can be compared with the red ones calculated assuming the CsCl would NOT sediment itself. There's a couple of clear differences:
1) The sedimentation boundary tends to move slightly faster (as compared to the homogeneous 'non-sedimenting' co-solute conditions) in the upper half of the solution column, because the co-solute is gradually being depleted from there. The boundary then slows down and moves slower, because of the co-solute accumulation here providing higher density (with some solvents also higher viscosity). The extent of this deceleration/acceleration pattern depends on the combination of s and D of the co-solute and the s-value of the protein.
2) The shape of the boundary is steeper than usual.
3) There's no flat 'plateau' region. The protein piles up toward the bottom due to the higher CsCl concentration increasing the buoyancy and decreasing the sedimentation rate. This is similar to the effect of solvent compressibility (3) - towards the bottom, the increasing density causes the flux into any volume element to be slightly larger than the flux out, therefore causing some continuous accumulation and sloping plateaus.
4) An additional inversion effect can show up just after the boundary: Because of the depletion of the co-solute from the meniscus region, the sedimentation here is faster than in the middle of the cell or at the bottom. Therefore, the macromolecules 'pile up' towards the middle of the cell (somewhere in the middle is the hinge-point for the co-solute, i.e. the point of little change in solvent properties). Note that this effect also will depend on the relative sedimentation rate of the protein and the co-solute. It is gravitationally stable because of the much larger co-solute concentration than macromolecule concentration.
Example 3: Experimental data of a protein in 1.4 M CsCl
In order to see if this effect of boundary inversion can be experimentally observed, we did an experiment with a ~ 80 kDa protein (5) in PBS with 1.43 M CsCl, at 60,000 rpm. Below are the data and the best-fit distributions, simulating the CsCl redistribution using the tabulated sedimentation properties of CsCl from (4). Water compressibility was considered in the model.
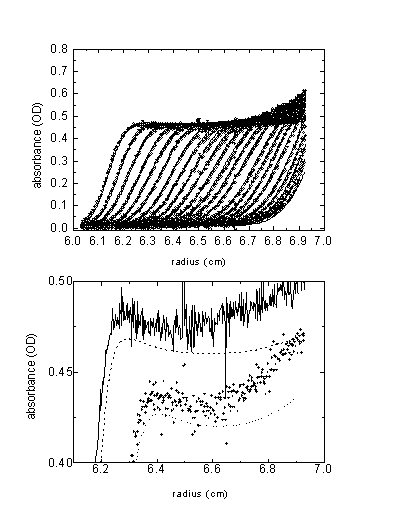
Overall, the features are consistent with the predicted ones. The boundary inversion is expected to be a small effect, but I think it was reproducibly visible. The lower plot shows two boundaries with the best-fit theoretical lines (offset for clarity). The best-fit apparent partial specific volume was 0.762 ml/g, indicating the preferential hydration of the protein in high CsCl concentration. For details, see (2).
Example 4: Analytical zone centrifugation with isopycnic conditions
The last illustration is the simulation in the configuration of analytical zone centrifugation: A solution of an 83 kDa protein (s = 5 S, v-bar = 0.77 ml/g) is layered on top of a solution of 2.5 M CsCl. Because there's no CsCl in the 1mm lamella on top, there will be initially a large density gradient at the boundary, which due to diffusion slowly approaches an equilibrium sedimentation profile of the co-solute similar to the homogeneous loading condition (see example 1). Shown below are the CsCl concentration distribution (molar units).
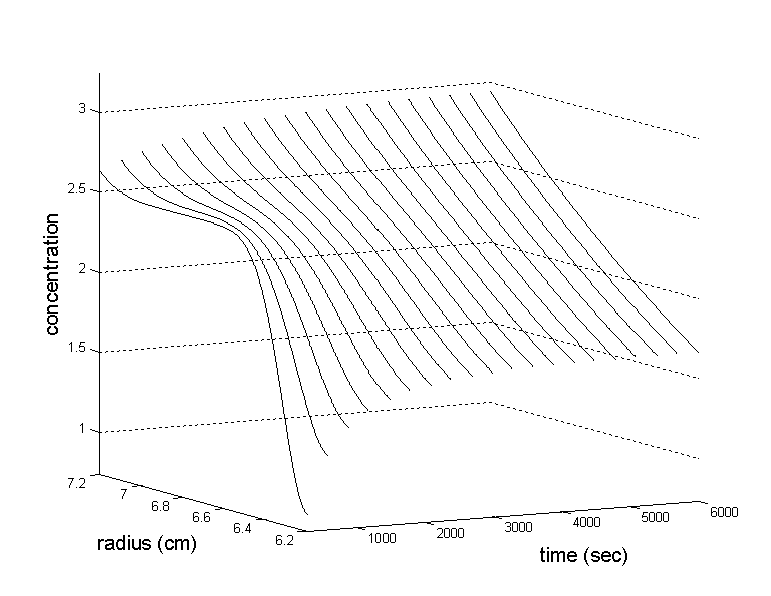
At the same time, the protein concentration distributions evolve as follows (times in multiples of 3000 sec, except the first distribution which is at 300 sec; starting distribution is shown as dotted line).
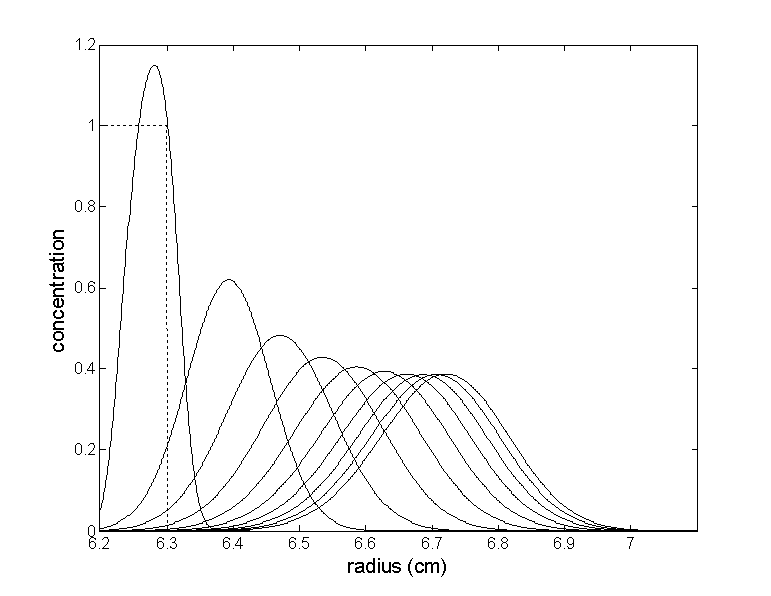
As you can see, the protein forms a lamella that moves down toward the point of neutral buoyancy. An interesting detail is that the protein initially reaches concentrations higher than the loading concentration. This is an effect of the protein piling up in the steep initial density gradient formed by the CsCl. Under some conditions, this effect can even produce a series of scans with initially increasing heights of the bands.
In order to describe the sedimentation of a macromolecule in an inhomogeneous solvent, we can use the Lamm equation
(Eq.
1) ![]()
but have to consider that the sedimentation and diffusion coefficients are not constant, but change locally due to the local solvent density. In general, this requires local corrections for solvent viscosity
(Eq.
2)
![]()
(with normal viscosity h20,w, local viscosity h(r,t)) and for solvent density
(Eq.
3)
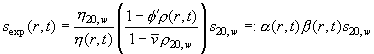
(with normal solvent density r20,w, local density r(r,t), normal partial-specific volume v-bar and apparent partial-specific volume F'). The corrections can be conveniently expressed with the factors a and b of relative viscosity and buoyancy, respectively. Constants are the diffusion coefficient D20,w and the sedimentation coefficient s20,w under normal conditions.
This approach neglects cross-diffusion terms in the general flux equations, which theoretically requires the absence of preferential hydration. However, in a first approximation we neglect those dynamic effects from the cross-diffusion terms (but not the preferential solvation effect for the buoyancy), assuming that the relative co-solute fluxes are small and that the preferential solvation is in a first order approximation does not change across the range of co-solute concentrations generated in the sedimentation process.
The problem of Eq. 1-3 is solved in a general way in ref 2. The strategy used takes the finite element Lamm equation method described in the Lamm equation tutorial, but extends it to expand the factors a and b also in terms of combination of elements Pk(r,t)
(Eq.
4)
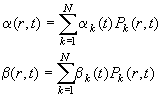
The additional terms from the expansion Eq. 10 cause the matrix elements A(1) and A(2) to become sums over tensor elements
(Eq.
11a)
![]()
(Eq.
11b)
![]()
, which leads us to the new matrices A(1*) and A(2*). Analytical expressions for the tensors are derived in ref 2.
The problem remains of determining the relative viscosity and buoyancy coefficients a(r,t) and b(r,t). This is done through a separate finite element simulation of the co-solute re-distribution. The time-steps of the co-solvent simulation are synchronized with the macromolecular Lamm equation solution. Sedimentation parameters for the co-solvent can be taken from tabulated values of s, D, and activity coefficients (2,4), or may simply be determined experimentally by modeling of interference optical sedimentation data. Currently implemented in Sedfit are ideal and non-ideal sedimentation for a single co-solute component. Having the local co-solute concentration, tabulated coefficients (such as from Sednterp) are used to calculate the local solvent density and viscosity.
References
(1) M. Meselson and F.W. Stahl (1958) The replication of DNA in Escherichia coli. PNAS 44:671
(2) P. Schuck (2003) A model for sedimentation in inhomogeneous media. I. Dynamic density gradients from sedimenting co-solutes. Biophysical Chemistry 108:187-200
(3) P. Schuck (2003) A model for sedimentation in inhomogeneous media. II. Compressibility of aqueous and organic solvents. Biophysical Chemistry 108:201-214
(4) A.P. Minton (1992) Simulation of the time-course of macromolecular separations in an ultracentrifuge. I. Formation of a cesium chloride density gradient at 25C. Biophys. Chem 42:13-21
(5) M.J. Rosovitz, P. Schuck, M. Varughese, A.P. Chopra, Y. Singh, L.J. McGinnis, and S.H. Leppla. (2003) Alanine scanning mutations in anthrax toxin protective antigen domain 4 reveal residues important for binding to the cellular receptor and to a neutralizing monoclonal antibody. Journal of Biological Chemistry (papers in press)
(6) R. Bruner and J. Vinograd. (1965) The evaluation of standard sedimentation coefficients of sodium RNA and sodium DNA from sedimentation velocity data in concentrated NaCl and CsCl solutions. BBA 108:18-29
(7) C.R. McEwen (1967) Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem. 20:114-149
(8) R.W. Clark (1976) Calculation of s20,w values using ultracentrifuge sedimentation data from linear sucrose gradients, an improved, simplified method. BBA 428:269-274
(9) M. Dishon, G.H. Weiss, and D. Yphantis (1976) Kinetics of sedimentation in a density gradient. Biopolymers 10:2095-2111
(10) W.K. Sartory, H.B. Halsall, and J.P. Breillatt (1976) Simulation of gradient and band propagation in the centrifuge. Biophys. Chem. 5:107-135