
back to the tutorial main page
Appendix:
Refine the analysis by exporting to Sedphat.
A quick overview of molar mass values:
After calculating c(s), we can use the function "Show Peak Mw in c(s)" to automatically print out molar mass estimates for each peak.

This is information is similar to the transformation of c(s) to c(M) described in the following. Accordingly, the molar mass estimate "Mf" is based on the best-fit frictional ratio (or other relationships for s and D if a specialized variant of c(s) is used). The value in parenthesis is the minimum molar mass of a spherical particle of given density.
As described above, these are clickable boxes that produce more information on each peak and how it relates to the raw data.
Conversion of c(s) to a molar mass distribution c(M)
After calculating the c(s) distribution and optimizing the parameters for the frictional ratio and meniscus, we can transform the c(s) distribution into a c(M) distribution. This can be done because in the calculation of c(s), for each s-value a diffusion coefficient D(s) was estimated (with the s-D relationship based on the optimal weight-average frictional ratio). Since s and D directly determines M, we also have M for each species, and in this way we can directly convert c(s) into c(M).

This leads to
which we need to rescale by drawing a rectangle with the right mouse button so as to show us the peaks:

resulting in
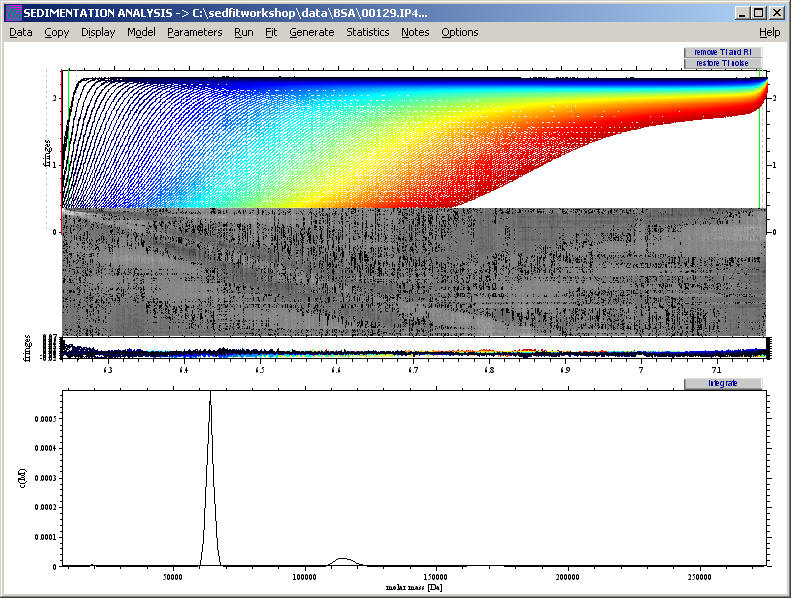
If we export the distribution again (using the copy command) into Origin, we can generate the following graph, which shows the oligomers of BSA with reasonable estimates of the molar mass:
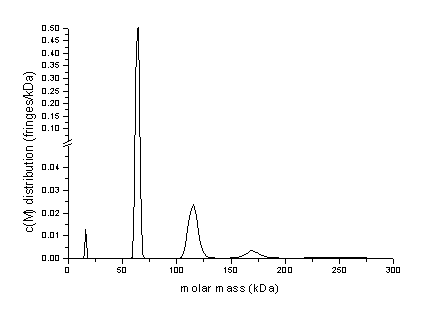
This may in general not work as well if the frictional ratio of the monomer differs much from the frictional ratios of the oligomers. In this case, the c(M) distribution will appear distorted.
New in version 8.7: It is sometimes possible, in particular if there is a substantial amount of oligomeric material with well-separated s-values, to use the model of a bi-modal c(s) distribution and to optimize two separate f/f0 values. For example, one could be the monomer, one for the higher oligomers. This is also useful if there are two different macromolecular compounds with widely differing s-ranges.
The best way to treat oligomerizing system is the SEDPHAT model for hybrid discrete/continuous distributions, in which the molar mass ratio of the oligomers can be constraint to be multiples. To get started with this, use the Export function of SEDFIT to transfer your data into SEDPHAT.
Further information, and comparison with other models can be found (here) .