This menu function will permit access to the local concentration parameters only for each experiment. There are other parameters which are local to each experiment, too, but are related more with the physical configuration of the experiment, and specified in the Experimental Parameters box. For more information on the different parameter types see the concepts for getting started.
The concentration parameters will depend on the particular model selected and on the experiment type. For non-interacting models, the concentration parameters are entered usually in the global parameters box, unless they are linear parameters, in which case no user input is needed (they are directly optimized in a single direct algebraic calculation). In both cases, the local parameter menu will not do anything. An exception is the species analysis with mass balance constraints.
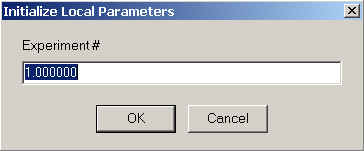
For the self-association model, you can specify the experiment number,

, specify if the local concentration parameter should be re-directed to that of another experiment (see introduction to shared local parameters)
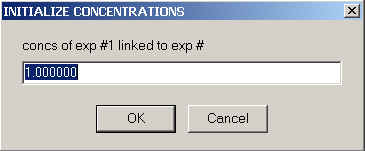
and (unless re-directed) enter the concentration in micromolar units.

For multi-speed sedimentation equilibrium experiments without mass conservation, you will have to enter the local concentration at the bottom of the cell (the bottom is the reference radius) for each scan. This is not required with the mass conservation option.
In heterogeneous associations, you will have to enter analogously the local concentrations and linkage of both components A and B. Heterogeneous associations can also be switched to concentration units of molar concentration of component A and molar ratios B/A.
Other information can be found in the specific models.
The local parameters menu will not be available for the non-interacting species models where the species concentrations are linear parameters (therefore no starting guess is necessary).
For the association models in the presence of a non-participating species, the local parameter menu expands, and the concentration of the non-participating species can be specified in signal units.
