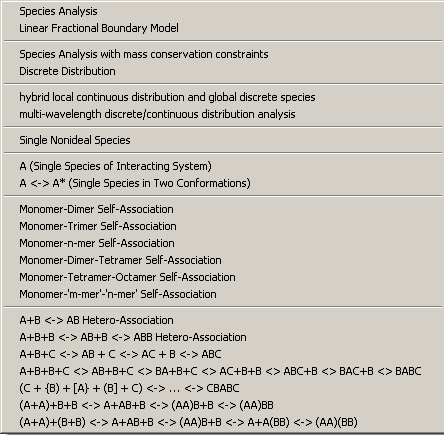

These are the models for global analysis. The list of models is planned to be continuously extended. (If you don't find the model you need, you could inquire for advance sharing of newer SedphaT versions with extended models - even if it doesn't exist, feedback on what users would like to have could be useful for me.)
Note that you can modify the default model SedphaT starts up with, and also for each model the default parameters. Further, you can store the global parameters of the models into files. These are *.sedphat_PAR files. They can be loaded or imported by drag-and-drop and applied to the currently loaded data. For more details, see Global Parameters, and the Data->Model functions.
![]()
Generally, you can distinguish the models in three major classes: non-interacting macromolecular species and mixtures, self-associating protein systems, and hetero-association systems.
The non-interacting models come in greater variety in global modeling than in the single experiment modeling of SedfiT. The reason is the different assumptions made as to whether the partial concentrations of the different species may be different, or if they are identical in the different experiments. First, one could imagine that because of different sample concentrations, the fractional concentration of a particular component of a mixture could be differently populated. Likewise, different populations may simply show up with higher or lower signal contributions in the different experiments. An example of the latter could be labeled mixtures with variable labeling efficiency for the different experiments, or dynamic light scattering experiments where the signal contribution is proportional to the molar mass of the particles. Second, it is possible that although all the fractional concentrations of all sub-populations are the same in different experiments, the total loading concentration might be different. This could be the case, for example, if different experiment employ different dilutions of the otherwise identical mixture. Third, a difference may occur in sedimentation equilibrium analysis as to whether mass conservation can be assumed or not.
Global modeling of mixtures of non-interacting species can be very useful, for example, combining sedimentation equilibrium data, sedimentation velocity data at different rotor speeds, and/or dynamic light scattering data.
The model of the sedimentation coefficient distribution of non-interacting species c(s) can also be applied to rapidly interacting species, if the peaks are interpreted in the framework of asymptotic boundary shapes in Gilbert-Jenkins theory. In this way, isotherms for the reaction boundary can be derived in this way.
Further, the model of component sedimentation coefficient distribution ck(s) can enable the determination of the composition of complexes formed by interacting proteins.
![]()
For sedimentation velocity data, these models include models for both slow chemical kinetics, as well as rapid chemical kinetics on the time-scale of sedimentation. They are straightforward, although the two-step association models are not implemented yet. No self-association model is implemented yet for dynamic light scattering.
Self-association models are studied best in a global analysis of several experiments conducted at a range of protein concentrations.
![]()
Heterogeneous association models
Models for rapid chemical equilibrium as well as slow chemical reactions are available. The more complex mixed association models are currently available only for sedimentation equilibrium. Note the comments on how to deal with the partial-specific volume for heterogeneous associations.
![]()
Note from version 4.0 and up: The appearance of the parameter input boxes for interacting systems will depend on the data type loaded. The goal is to simplify the parameter entry by eliminating irrelevant parameters . For example, enthalpy number are only required in ITC experiments, and kinetic rate constants currently only for sedimentation velocity.
Follow these links for more detailed information on the particular models:
![]()