A number of different isotherm types can be loaded in SedphaT. Here is a general overview of their use.
Note that several isotherms can be loaded simultaneously and analyzed globally. For example, from a single series of sedimentation velocity experiments at different loading composition, isotherms of the weight-average s-value, populations isotherms, and for rapidly reacting systems Gilbert reaction boundary isotherms can be obtained and analyzed globally. This can make use of different aspects of the sedimentation data.
The following is biased towards sedimentation velocity experiments, but isotherms from other experiments, such as sedimentation equilibrium can also be used. In particular, any technique that provides a signal that is a weighted population average can be analyzed in the same form as weight-average s-values. Just replace the 's-values' of each species with the characteristic observables.
(See other pages for detail on the formatting requirements and the experimental parameter box. The choice of concentration pairs of A and B in the isotherm can be arbitrary, but if no system is recognized, each data point will have one fitted line (representing slices through the cA-cB-surface)).
Weight-average s-values and Mw values
This extracts thermodynamic information from sedimentation experiments. For sedimentation velocity, this is completely independent on boundary shape and valid for any reaction kinetics. It is based solely on considerations of mass balance, and simply requires that the model used to extract the sw value from the sedimentation profiles empirically fits the sedimentation boundary well.
The most precise way to determine sw is by doing a c(s) analysis and integrating the peaks over all peaks from species participating in the interaction.
A detailed analysis can be found in
P. Schuck (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Analytical Biochemistry 320:104-124
c(s) is described here, the integration tool here.
Assessing the reaction kinetics
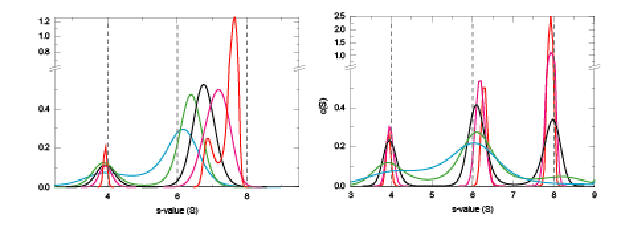
More detailed information can be obtained exploiting the hydrodynamic separation of the sedimenting species. This will depend on the reaction kinetics, which can be assessed by a c(s) analysis. As described in the website on sedimentation velocity for reactive systems, for a heterogeneous association this can look like this:

On the left is shown a fast interaction on the time-scale of sedimentation, with the typical undisturbed boundary and the concentration-dependent reaction boundary, and on the right hand side is shown a slow interaction (under otherwise identical conditions).
Which isotherm analysis to choose depends on the time-scale of the interaction:
partial-conc. isotherm: for slow interacting systems where the c(s) peak positions remain independent of loading composition. Data are amplitudes of c(s), determined by integration of c(s) peaks in SedfiT.
Gilbert sw fast: for fast interacting systems, which can be discerned from c(s) peaks showing peaks positions depending on loading composition. Data are the weight-average s-value of the fast boundary component in c(s), as determined by integration in SedfiT.
Gilbert partial conc: for fast interacting systems, which can be discerned from c(s) peaks showing peaks positions depending on loading composition. Data are the amplitudes of both the undisturbed boundary and the reaction boundary component in c(s), as determined by integration in SedfiT.
These isotherms are described in detail in
J. Dam, C.A. Velikovsky, R.A. Mariuzza, C. Urbanke, P. Schuck (2005) Sedimenetation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophysical Journal 89:619-634
J. Dam, P. Schuck (2005) Sedimentation velocity analysis of heterogeneous protein-protein interactions: Sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophysical Journal 89:651-666
The isotherms based on Gilbert-Jenkins theory are new in version 3.0. They exploit the boundary structure of reactive systems, and the fact that c(s) can be understood as an approximation of the asymptotic reaction boundaries at infinite time (for more information, see this presentation: c(s) based analysis of rapidly interacting systems).
Note that these isotherms can be fitted globally with the weight-average s-values to give optimal results. The raw asymptotic boundaries as predicted by Gilbert-Jenkins theory can be saved using the "save intermediate results" option in the Lamm equation parameter box.
New in version 2.0
Sedimentation velocity simulations for reacting systems have been added. This includes self-associating systems, as well as hetero-associating systems both with finite reaction kinetics and with instantaneous equilibrium (on the time-scale of sedimentation).
These models can be used to directly model the sedimentation boundary for reacting systems. It is an alternative to the analysis of the analysis of the isotherm of weight-average s-values. The boundary modeling extracts more information from the sedimentation data, specifically on the reaction kinetics.
The reaction kinetics model simulates the chemical conversion kinetics of the interacting species during the sedimentation process. For systems with off-rate constants > 0.01/sec, the reaction can be considered essentially instantaneous on the time-scale of sedimentation under most conditions (exceptions are theoretically possible for extremely large molecules sedimented at extremely high rotor speed). Therefore the sedimentation profiles become nearly identical to those calculated under conditions where the mass action law is fulfilled instantaneously at all times throughout the cell. This second case limiting is also implemented in Sedphat independently from the reaction kinetics model. [The instantaneous mass action law model for self-associations is a special case of the Lamm equation with locally weight-average s-values and gradient-average D-values, but no such relationship exists for heterogeneous associations.]