time-dependent s and M
Model | Special | Fit time-dependent s, M (e.g. in Archibald Analysis)
Sometimes it can be useful to allow for (apparent) changes in M and s with time. In Archibald-type analyses, for example, where only the initial depletion at the meniscus is analyzed in the context of a Lamm equation model, heterogeneity can be detected by a drop of M and s with time, because the larger species are depleted more rapidly, therefore lowering the average observed in the vicinity of the meniscus (ref 1).
New in version 8.7: Note the new (differential) second moment methods that can also reveal sw(t) data.
While a normal Archibald-type analysis can be performed by a straight-forward application of the model for noninteracting discrete species, analyses of the time-dependence can be done with this special function. The idea for analyzing the time-dependence is that in a sequence of analyses, different subsets of the available scans are considered, either in a moving time-window approach, or by continually increasing the time-window.
This procedure consists of the following steps:
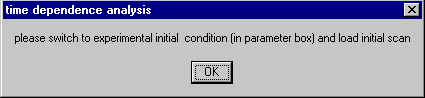
1) The analysis mode needs to be switched to ‘experimental initial conditions’ in the parameters box, and an initial scan - most likely the first available scan - must be loaded (see load initial data and extrapolation of initial data). A message box will remind of this step:

(Select the noninteracting discrete species model, click on the parameter menu and switch to the experimental initial distribution. A warning message may appear that reminds that the rotor acceleration corrections are switched off; this can be ignored here. )
2) Select Model | Special | Fit time-dependent s, M. The number of scans n that are pooled in the time-dependence analysis must be specified:

The best value here will depend on the number of scans available, the desired resolution in time, and the level of noise in the scans. Usually, the best number can be empirically determined. It should not be too small, though, because this increases the noise and decreases the stability of the procedure.
3) The choice of a moving average 1 to n, 2 to (n+1), 3 to (n+2),... or an increasing window 1 to n, 1 to (n+1), 1 to (n+2), etc. appears.
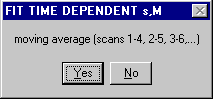
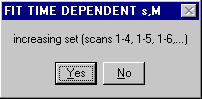
Because of the increasing number of scans in the second option, this is numerically more stable and leads to slightly higher precision, although smaller changes with time.
4) A series of fits is made, each to a subset of the scans.
Please Note:
Sometimes during the course of this series of fits, bad parameters are encountered in the Simplex routines. If the gridsize is slightly reduced (e.g. to 250), and if the number of scans per set is increased, this can improve the stability of the method.
This procedure has not been fully optimized. In my hands, the increasing window gives generally better results.
5) The results are stored in two files: mwdcdt.dat and sdcdt.dat (if the moving average was selected) or Mw(t).dat and s(t).dat (if the increasing time-window option is selected). Each file contains two-column ASCII text with the average times of the scan subsets considered, and the best-fit M and s, respectively. The files are located in the directory with the data.
Please Note
: This analysis works only with model for noninteracting discrete species.
References:
(1) P. Schuck and D.B. Millar. (1998) Rapid determination of molar mass in Archibald experiments by direct analysis using the Lamm equation. Analytical Biochemistry 259:48-53