Analytical Zone Centrifugation
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation Indep. Species
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation c(s)
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation ls-g*(s)
Analytical zone centrifugation is a technique in which a small volume of sample (typically 20 ml) is layered on top of solvent (e.g. 330 ml of solvent with slightly higher density), and the sedimentation process of the macromolecules is observed. For technical details on the experiment, see ref 1.
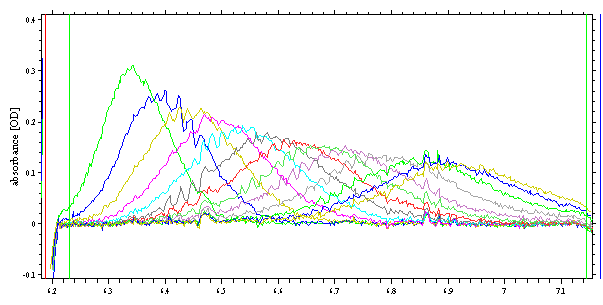
Typical data may look like Gaussians that move and broaden with time (these data are from an experiment with BSA).

The sedimentation process is described by the Lamm equation, just like ordinary sedimentation velocity experiments. The only difference is the starting condition: Usually, the sedimentation starts with macromolecular concentration uniform across the cell, while in the analytical zone centrifugation the starting condition is a small lamina of uniform concentration near the meniscus up to the interface, and then zero concentration from the interface to the cell bottom.
There are two different approaches for the quantitative analysis of these data by direct boundary modeling: First, one could chose the first scan as initial condition, and use the conventional Lamm equation model. In more detail, this can be done by using the Data | Set Initial Data command. As described on the help website of the experimental initial function, first select the model for non-interacting discrete species, then invoke the parameter box and switch on the experimental initial condition. Then load the initial data. The advantage of this method is that any imperfections in the overlaying process is irrelevant, as only the evolution after the time of the first scan will be modeled. Disadvantage is that this model can strictly be only used with a single species model, or with self-association models. (The reason is that for multiple macromolecular components, we would need separate initial conditions for all of them.)
As an alternative, we can assume that the overlaying process is ideal and instantaneous. This means that we can build into the Lamm equation solutions the initial condition of a homogeneously loaded layer of a certain thickness. This is the strategy the specialized analytical zone centrifugation models are for.
The lamina thickness will be an additional parameter, but we will be able to use any of the direct boundary sedimentation models. The lamina thickness (e.g. ~ 0.09 - 0.10 cm for 20 ml over 330 ml buffer) will introduce some correlation with the sedimentation or diffusion parameters. I would recommend to keep the lamina thickness a floating parameter in the analysis, together with the meniscus position.
It should be kept in mind that the solvent density and partial-specific volume of the macromolecules should be entered in SEDFIT, in particular if D2O was used in the solvent, the buoyancy of the molecules can be much different. (Also there is a large viscosity effect of D2O). For tabulated data, see ref 1 and ref 2.
Because of the difference in composition of the sample and the solvent required for formation of the diffusional density gradient, only absorbance optics can be used in this experiment.
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation Indep. Species
This model works like the model for independent and ideally sedimenting species for conventional loading. The parameter box is slightly different:
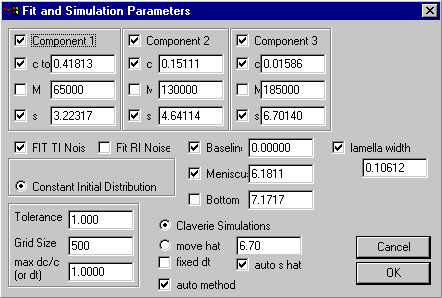
It allows only for 3 components instead of four, but it has an additional parameter for the lamella width. As mentioned above, I recommend keeping the lamella width a floating parameter (check the box left to "lamella width"), as well as the meniscus. For the BSA data shown above, we can fit the data well with the three main species:
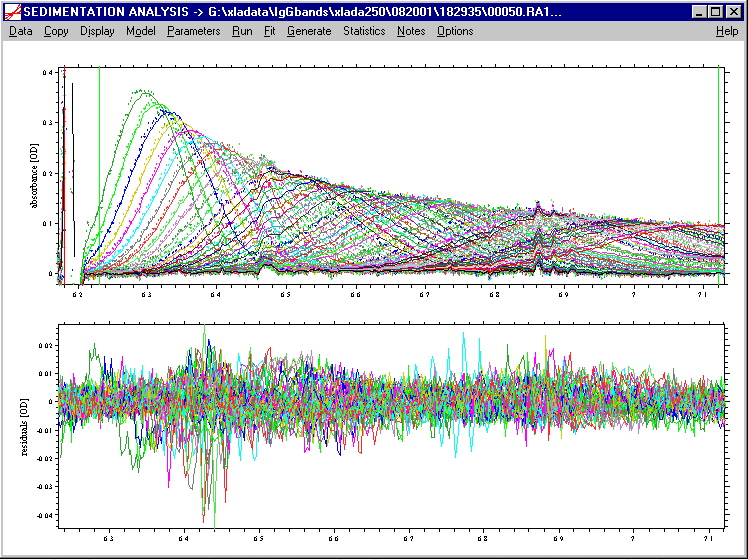
With consideration of TI noise (because of the clear dust spikes on the data), we get ~ 15% dimer, and 1-2% trimer, with a rmsd of the fit 0.0043 OD.
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation c(s)
This model is very analogous to the c(s) distribution for conventional loading, but there are some differences.
Because of the higher correlation, I recommend the use of the Tikhonov-Phillips regularization, and the use of a higher confidence level, such as 0.95. Otherwise, I suggest using the same optimization procedure as described in the step-by-step tutorial, with floating meniscus, lamella thickness, and frictional ratio.
The best-fit distribution might look like this:
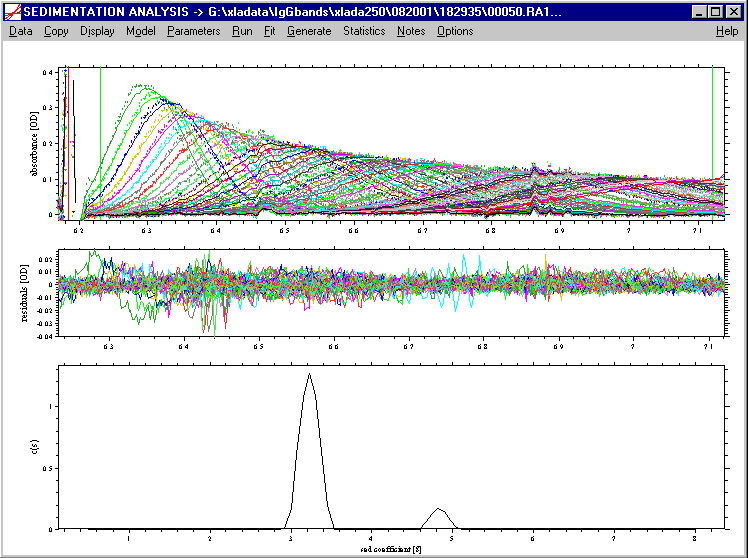
Model | Analytical Zone Centrifugation | Analytical Zone Centrifugation ls-g*(s)
Like the other models, also the ls-g*(s) model can be modified to take the initial layer into account. As described in the introduction to ls-g*(s), this model is based on step-functions that describe the sedimentation in the absence of diffusion. The modification required for analytical zone centrifugation is that the basic step-function is actually two steps (with concentrations zero - cplat - zero). Details of the model will be described elsewhere (ref 2).
The complete analysis can be done analogous to the ls-g*(s) model for conventional loading, similar to the procedure shown in the tutorial. The parameter box will have the additional parameter for the lamella width:
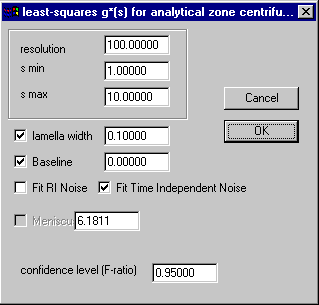
and fitting for the meniscus position is not implemented. I do not recommend fitting for the lamella width here, as there can be some slight correlation with the distribution shape.
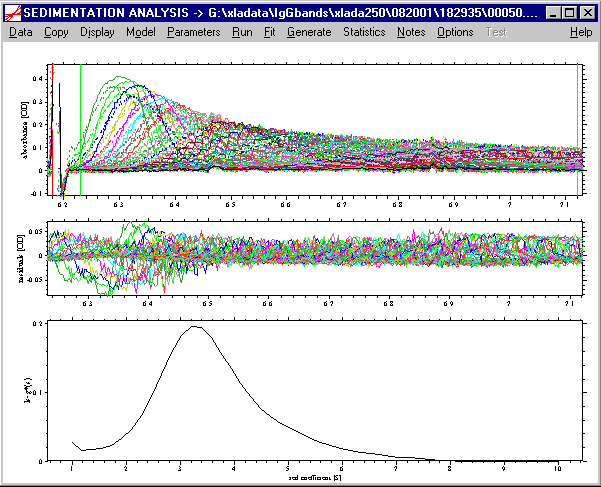
As described in the introduction to the ls-g*(s) method, we can use a very large data set, and the result may look like this:
References
(1) J. Lebowitz, M. Teale, P. Schuck (1998) Analytical band centrifugation of proteins and protein complexes. Biochem. Soc. Trans. 26:745-749
(2) manuscript in preparation