van Holde-Weischet analysis
Model | van Holde-Weischet
Model | extrapolate ls-g*(s) vs 1/sqrt(time)
An introduction to the theory and practice of the analysis method by van Holde and Weischet (ref 1) can be found here. Reading of this website is suggested before use of the method. As described in this introduction, it is an elegant extrapolation method for boundary diagnostics. For quantitative analysis of the sedimentation data, these method are surpassed in many respects by direct boundary models, especially by Lamm equation solutions and size-distributions c(s).
Very important for the use of the van Holde-Weischet method is the selection of appropriate data subsets. As explained in the tutorial, we need for each scan plateaus to be established, and we need the solvent plateau near the meniscus.
This model is only good for absorbance data.
The convention in Sedfit is that the plateau must be established for all scans within the 0.05 cm left to the outer fitting limit (take the right green line and go back 0.05 cm -- this should all be flat in all scans). The solvent plateau must be established within the 0.05 cm right to the inner fitting limit of the last scan (look at the left green line, go 0.05 cm to larger radii -- this should be flat for the last scan).
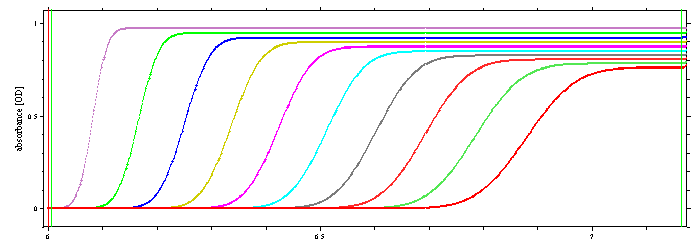
The following data set fulfills this condition:

When switching on the van Holde-Weischet model, a message box appears:

The analysis can be performed for sedimentation data and for flotation data. Usually, you can simply press Enter key here, unless when studying floatation processes (the analysis is the same, but SEDFIT must be told which one to use). In case of flotation, all the information about required plateaus is understood in reverse, i.e. solution plateau near the meniscus for all scans and solvent plateau near the bottom required for the last scan.
Next, the resolution N (i.e. the number of boundary divisions) must be entered:

By default, a value of 50 is recommended. This value is probably too high in most cases, because it leads to very narrow increments of boundary fractions. Dependent on the noise level of the data, there may be scans that actually do not have any data points in one or more of the boundary slices. If this is the case, the resolution will be automatically reduced, until every scan has at least one data point in all fractions. This will be a good value for the resolution, appropriate for the least-squares determination of the radial position of the boundary fractions (see the implementation).
Optionally, the well-known plot of the linear regression of the boundary fractions will be shown, if yes (or press Enter key)

Please note that the Parameters command does not apply here (there will be no action when clicking on this menu), because there are no further parameters to set. If the resolution parameter is to be changed, click with the mouse no the Model | van Holde-Weischet menu again.
The analysis will be executed by a Run command. If necessary, the number of boundary fractions will be reduced, indicated by a text output:
dividing data in .. boundary fractions
and finally a message box:
Press OK (or hit the Enter key), and two new graphs will appear instead of the data and residuals plot: In the upper plot the apparent sedimentation coefficient values for each boundary fraction versus inverse square root of time, and in the lower plot the integral sedimentation coefficient distribution G(s).

This s(app,i) plot and its data can be copied using the copy data plot and the copy data table command. (The copy data table command will give a table with 1/sqrt(t) in the first column, followed by the best-fit values for each fraction (one column is one fraction), followed by the experimental data for each fraction [ignore the 0 at the 1/sqrt(t) of 0]). The G(s) distribution can be exported using the copy distribution table command.
Please note that the scale in the G(s) plot is determined by the minimum and maximum s-value occurring. The above example, the distribution is actually very precise, as can be seen in a different axis scale (after exporting):

Pressing the ESCAPE key will return to the usual display of the data and residuals, and in the bottom part of the window, the integral distribution G(s).
It should be noted that here, Sedfit shows the best-fit position of each of the boundary fractions directly in the raw data space, i.e. it calculates backwards from the linear regression of s(app,i) vs 1/sqrt(time) to best-fit s(app) values, and best-fit positions of the boundary fractions. This has the advantage that it allows to judge how well the data are described by the van Holde-Weischet analysis.
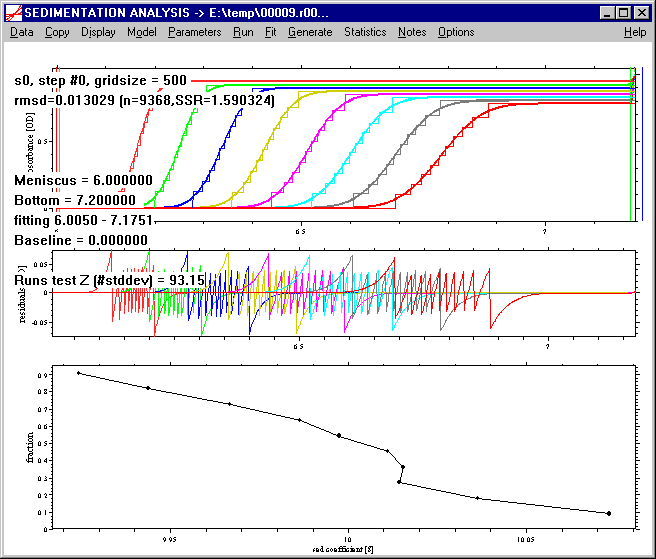
Troubleshooting:
If aberrant boundary fractions occur near the upper or lower end of the G(s) distribution, I recommend recalculation of the analysis (click with the mouse no the Model | van Holde-Weischet menu again) with a significantly lower resolution. (Aberrant fractions are usually a result of noise of the data being larger than the concentration increment of the boundary division. This averages out somewhat in the middle fractions, but noise from the plateaus reaching into close-by boundary fractions gives a one-sided bias.) Alternatively, if the resolution in the middle fractions shall not be sacrificed, the distribution can be exported, and the aberrant extreme fractions removed.
B) Model | extrapolate ls-g*(s) vs 1/sqrt(time)
In this variant, the position of the boundary fractions are not obtained directly from the data. Instead, apparent s-values for boundary fractions are obtained via equivalent area fractions of a series of ls-g*(s) curves, which then are subjected to the same 1/sqrt(time) extrapolation. This allows working in the presence of time-invariant and radial invariant noise. This model is presented in more detail in the tutorial.
Because of a constant total area under all the ls-g*(s) curves is needed, similar constraints on the selection of data sets are imposed. To avoid correlation of the baseline parameters with small s-value part of ls-g*(s), all scans should be well clear of the meniscus. For a well-determined high s-value limit of ls-g*(s), all scans should have a clearly established plateau.
It was found to work best when RI noise has been eliminated, for example by manual removal of jitter, and switching of RI noise in the parameter box. (This does not apply to the TI noise, as TI noise can be taken into account.) This model can be applied equally to absorbance or interference data.
After selecting this model, a warning message appears,
as a reminder to the experimental character this model still has. Although very good results have been obtained (ref 2), overall very little experience with this approach is available. Further, the extrapolation of ls-g*(s) has the same fundamental limitations in resolution and sensitivity as the van Holde-Weischet method (see the limitations), and is surpassed by the c(s) model.
After selecting the model, a message box

appears, similar to the van Holde-Weischet model. However, more input is required for calculating ls-g*(s) which can be done by clicking on the Parameters menu:

The top portion of the parameters box determines the ls-g*(s) parameters (familiarity with the ls-g*(s) model and the tutorial is required). The fields for resolution ('gofs resolution'), s-min, s-max, and confidence level are the same as in the other size-distribution models. A relatively low value for the gofs resolution (50) can be sufficient.
Because we need to calculate ls-g*(s) curves at different times, the total number of scans is subdivided in subsets, which are input in the field "#scans for g*(s)". Some care has to be taken that this number is not too small, resulting in instable ls-g*(s) curves, and not too large, resulting in poor time resolution. I have good experience with a value of 10, but since I have not systematically optimized this to the stage of developing a general rule for good default values, some testing with different numbers may have to be done.
Connected to this is the way in which the sets of scans are made, either by combining the scans in sequences like [1,2,3] [4,5,6], etc. or like a moving box [1,2,3], [2,3,4], [3,4,5], etc. Check ‘slide frame’ box for this second method, leave it unchecked for the first. Obviously, the sliding frame has the advantage of providing more ls-g*(s) curves, but it also will take much more time.
The middle part is concerned with the parameters for the extrapolation of the area (corresponding to the boundary) fractions. The only parameter required is the number of boundary divisions. I recommend here a not too high number (< 50), although due to the smoothness of the ls-g*(s) distribution a larger number can be used than in the van Holde-Weischet method.
The lower part, finally, consists of the usual baseline parameters and the meniscus position. Please note that it is possible to include systematic noise in this model, because each of the ls-g*(s) can be calculated considering systematic time-invariant or radial-invariant noise.
When the parameters have been specified, click on the Run command to start the analysis. This will cause the series of ls-g*(s) distributions to be calculated. They are shown superimposed, and it can be observed how the g*(s) distribution gets sharper with time (i.e. for the sets of later scans). The result is shown in a similar representation as in the van Holde-Weischet analysis:
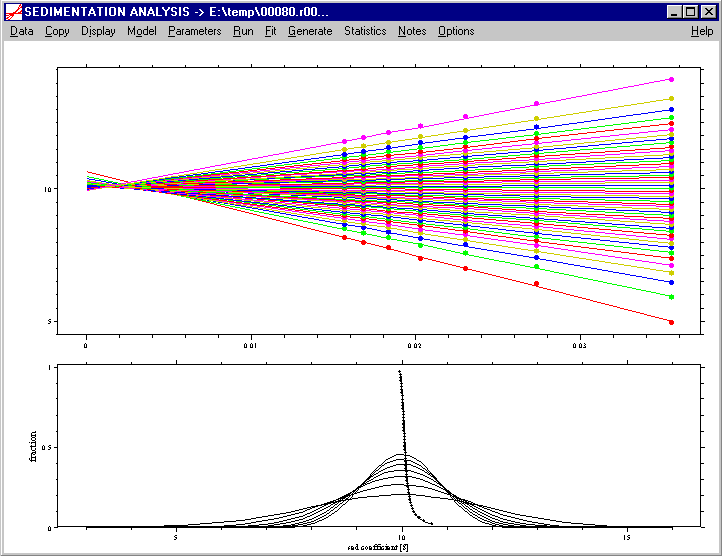
The extrapolation of the area fractions of ls-g*(s) on top, and the original ls-g*(s) with the extrapolation G(s) in the lower graphics. Again, they can be copied using the copy data plot and the copy data table command. The G(s) distribution and the ls-g*(s) distributions can be exported using the copy distribution table command. When using this copy distribution table command, a message box appears
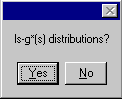
prompting for a selection if either the complete set of ls-g*(s) or the G(s) distribution should be placed into the clipboard.
Pressing the ESCAPE key will return to the usual display of the data and residuals, and in the bottom part of the window, the integral distribution G(s) together with the ls-g*(s) distributions.

Similar to the van Holde-Weischet analysis, from the linear regression of the apparent sedimentation coefficients s(app,i) of the boundary/area fractions, we can get the best-fit s(app,i) values, and as a consequence, the best-fit radial positions of the boundary fractions. These best-fit boundary fractions can be composed into an explicit boundary model, which allows calculating the overall best-fit systematic noise contributions.
Some experimenting with optimal parameters may be required, as the G(s) distribution can be sensitive to outlier g*(s) distributions. It seems advantageous if the systematic noise can be reduced to either RI noise or TI noise, as I have observed that both together can lead to poor extrapolation in the lower boundary fractions.
References
(1) K.E. van Holde and W.O. Weischet. (1978) Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers 17:1387-1403
(2) P. Schuck, M.S. Perugini, N.R. Gonzales, G.J. Howlett, and D. Schubert. (2002) Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophysical Journal 82:1096-1111