[download] [getting
started] [examples] [tutorials]
[sedfit help] [FAQs]
[references] [links
to other AUC sites] [contact] [
Search ] [
sedphat ]
Sedfit
This website provides information on Sedfit, a software
for the analysis of analytical ultracentrifugation and other hydrodynamic data,
written at the National Institutes of Health and distributed without charge for
research use. The program can be downloaded from
here, and it is also available directly from
sedfitsedphat.nibib.nih.gov, or it can be sent as an email attachment from
the author on request).
As documentation please refer to the following books, which have specific
information on SEDFIT and SEDPHAT functions as side-bars nested into the
description of theory and experiment of analytical ultracentrifugation (AUC):
https://www.crcpress.com/Basic-Principles-of-Analytical-Ultracentrifugation/Schuck-Zhao-Brautigam-Ghirlando/p/book/9781498751155
https://www.crcpress.com/Sedimentation-Velocity-Analytical-Ultracentrifugation-Discrete-Species/Schuck/p/book/9781498768948
https://www.crcpress.com/Sedimentation-Velocity-Analytical-Ultracentrifugation-Interacting-Systems/Schuck-Zhao/p/book/9781138035287
A general introduction to the study of protein interactions by analytical
ultracentrifugation can be found at the website of our lab at NIH (DMAS/LCIMB/NIBIB).
This includes an introduction to the general principles of AUC and detailed
experimental
protocols.
The main
purpose of this website is to provide additional information for using
the software, such as an online help reference information,
and background information (tutorials) on direct boundary modeling, FAQs,
references,
examples and more.
for
upcoming workshops: see
https://sedfitsedphat.nibib.nih.gov/workshop/default.aspx
Please join the
Sedfit Users Group
email list (reaches currently > 900 AUC users) for technical advice and discussions with other users on software
issues and applications. You can also sign up to the
SEDPHAT-L listserv for discussion of applications of SEDPHAT and questions
of global analysis of ITC, AUC, DLS, SPR, and other biophysical techniques.
The main features of Sedfit include:
 | analysis of data from: sedimentation velocity, dynamic light
scattering, and sedimentation equilibrium.
Sedimentation velocity data can be acquired using absorbance or interference optics,
in conventional loading configuration or synthetic boundary, analytical zone
centrifugation, and different cell geometries |
 | continuous size-distributions c(s) with many variants of prior knowledge for sedimentation velocity
analysis with maximum entropy regularization |
 | size-and-shape distributions c(s,M) |
 | Bayesian incorporation of prior for enhanced resolution of distributions |
 | Lamm equation models for: discrete non-interacting species,
self-associating systems (1-2, 1-3, 1-2-4, 1-4-8, 1-m-n), non-ideal sedimentation
--> see extension for global analysis of interacting systems in
Sedphat
|
 | apparent sedimentation coefficient distribution ls-g*(s) and van
Holde-Weischet analysis G(s) (both for absorbance and interference data) |
 | all sedimentation velocity models for
direct boundary modeling with algebraic noise elimination |
 | corrections for water compressibility (and compressible organic solvents) |
 | continuous size-distribution models for dynamic light scattering and
sedimentation equilibrium |
 |
numerous statistical functions, loading options, and general purpose tools |
 |
extension to global analysis in
Sedphat
|
Sedfit
is closely related to Sedphat,
which provides global modeling capability for both sedimentation equilibrium
and sedimentation velocity data. It also can serve as a platform for
the global analysis of a variety of isotherms from different biophysical
disciplines. 
NEW:
 | accounts for finite time of absorbance scanning |
 | Extended multi-threading support for faster analysis on computers
with multi-core processors |
 | Bayesian prior knowledge for enhanced resolution of c(s) (links
to
paper and
how to use in SEDFIT) |
 | faster Lamm equation solutions (description) |
 | new optimization tools: Marquardt-Levenberg and simulated annealing
(how to use in
SEDFIT) |
 | real-time testing for attainment of sedimentation equilibrium (how
to use in SEDFIT) |
 |
multi-thread support |
 | new user interface (new color scheme, wizard messages, graphical
editing of data points and scans, dragging limits with the mouse,
quick change of cells,
pre-set limits for automatic integration) |
 | version 9.4: new specialized c(s) models (wormlike
chain,
floating vbar), extended calculator,
display, and other
utility functions (improved 6-channel data support) |
 | version 9.3: size-and-shape distributions c(s,f) and c(s,M),
and the scale-relationship free general c(s,*) |
 | sounds,
serialization of analysis (apply the same analysis automatically
to data from several cells) |
 | version 9.2: optional speeding up of c(s) analyses by slight
radial
pre-averaging of scans, faster
Lamm equation
solutions with permeable bottom model,
c(s) model
with user-defined M-s relationship,
optional
updating of display while fiitting, restoring previous c(s) analyses,
loading data via drag-and-drop, importing SEDPHAT xp-files, and other new and
improved utility functions. |
 | new tutorials on solvent compressibility,
and self-forming
density gradients |
 | new Sedphat hybrid
bimodal c(s)-discrete species model combining 2 c(s) distributions with
discrete species, for example allowing to link molar mass into integral
multiples |
 | version 8.9: Export data to Sedphat,
and spawn Sedphat
automatically with current data (and c(s) hybrid model). |
 | new in version 8.8 (03/04): maintenance update with a few bug fixes and
added flexibility |
 | new in version 8.7 (09/03): sedimentation in inhomogeneous
solvents, like sedimenting co-solutes and compressible solvents,
including corrections for water
compressibility;
extended Monte-Carlo error analysis for weight-average s-values or
concentrations of trace components;
differential second moment method for sw; faster c(s)
fitting; improved ellipsoid shape calculator; bimodal
c(s) with two f/f0 parameters, back-transform for g(s*), several new utilities and bug fixes |
 | new mode for REALTIME analysis |
NEW
REFERENCES:
 | Bayesian analysis of trace components (AAPS
Journal 10 (2008) 481-493) |
 | Bayesian prior knowledge (Biomacromolecules
8 (2007) 2011-2024) |
 | new Lamm equation solutions (Computer
Physics Communications in press) |
 | size-and-shape distributions c(s,M) and scale-free general c(s,*) (Biophysical
Journal 90 (2006) 4651-4661 full text ) |
 | interpreting c(s) for reacting systems:
Biophysical Journal
89:651-666 and
Biophysical Journal
89:619-634 |
 | tutorial
review of analytical ultracentrifugation in protein science (Protein Science 11 (2002) 2067-2079) |
 | new review of the strategies and numerical methods for sedimentation
coefficient distributions in Methods
in Enzymology (in press) |
 | studies of strategies for using sedimentation velocity for protein
self-association: Analytical
Biochemistry 320:104-124 |
 | macromolecular sedimentation in the presence of sedimenting
co-solutes (dynamic density gradients): Biophysical
Chemistry 108:187-200 |
 | corrections for solvent compressibility Biophysical
Chemistry 108:201-214 |
A snapshot from the direct boundary modeling of data from a sample of IgG,
analyzed with a c(s) distribution of Lamm equation solutions illustrates the
rich information contained in sedimentation velocity data.
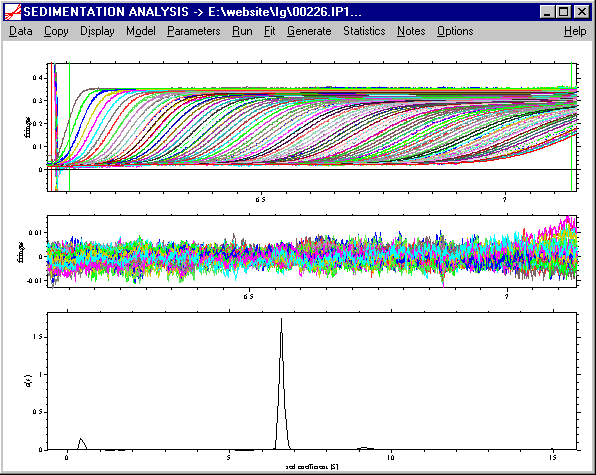
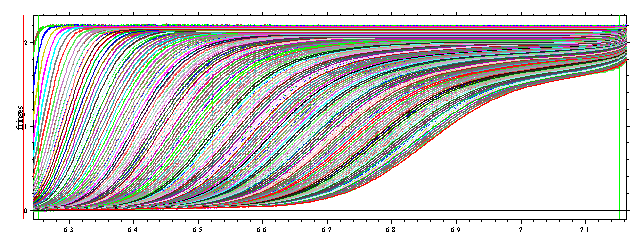
This example also illustrates some of the main ideas of Sedfit:
loading data from the entire sedimentation process, use of systematic noise
decomposition (and subtraction), modeling with finite element solutions of the
Lamm equation. If we expand the scale of the continuous sedimentation
distribution c(s) with maximum entropy regularization shown above, it can be
seen that the c(s) analysis reveals the presence of
several oligomeric species and a smaller species:
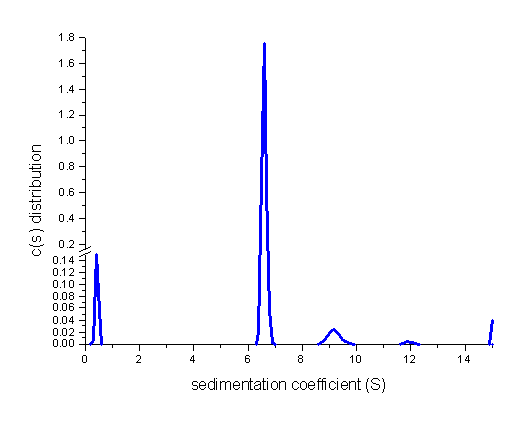
Similarly, the analysis of data from a BSA sample
shows the presence of dimer and trimer in the
sedimentation coefficient distribution c(s)
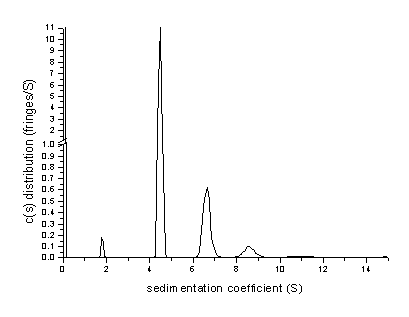
Transformation to a molar mass distribution (assuming that all species have a
similar frictional ratio) is consistent with the oligomeric BSA
species:
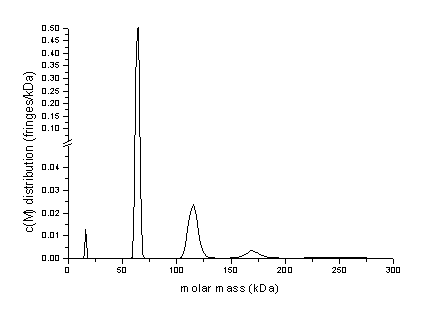
Analyses free of scale-relation ship assumptions of similar frictional
ratios are available, such as the two-dimensional
size-and-shape distribution c(s,f),
c(s,M), and the general c(s,*).
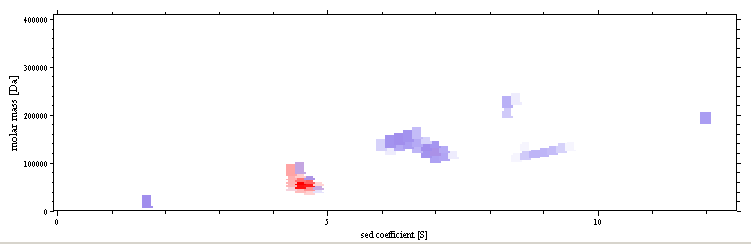
Sedfit offers several different
options for utilizing different prior knowledge on the sample under study (such as the
number of discrete species, a self-association model, etc.). More
information on these examples can be found in the step-by-step
tutorial (BSA) and the example for using Sedfit
(IgG).
For questions please contact the
Sedfit Users Group
email list .
DISCLAIMER STATEMENT:
The information contained herein is provided as a service
with the understanding that the author makes no warranties, either expressed or
implied, concerning the accuracy, completeness, reliability, or suitability of
the information.
[download] [getting
started] [examples] [tutorials]
[sedfit help] [FAQs]
[references] [links
to other AUC sites] [contact]
![]()





